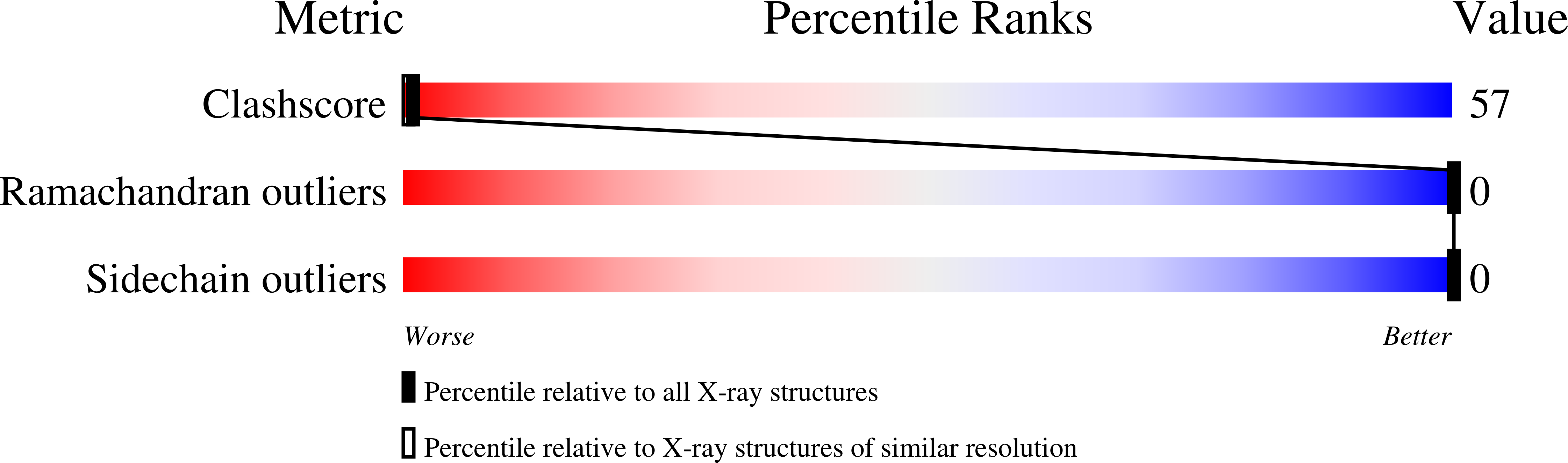Nonstoichiometric Complex of Gramicidin D with Ki at 0.80 A Resolution.
Olczak, A., Glowka, M.L., Szczesio, M., Bojarska, J., Duax, W.L., Burkhart, B.M., Wawrzak, Z.(2007) Acta Crystallogr D Biol Crystallogr 63: 319
- PubMed: 17327669
- DOI: https://doi.org/10.1107/S0907444906053649
- Primary Citation of Related Structures:
2IZQ - PubMed Abstract:
The crystal structure of a nonstoichiometric complex of gramicidin D (gD) with KI has been determined at 100 K using synchrotron radiation. The final R factor was 0.106 for 83 988 observed reflections (Friedel pairs were not merged) collected to 0.80 A. The structure consists of four independent pentadecapeptides and numerous solvent molecules and salt ions. The general architecture of the antiparallel double-stranded gramicidin dimers in the crystal (a right-handed antiparallel DSbetaH(R) form) closely resembles that of previously published cation complexes of gD. However, a significantly different mixture of gramicidin isomers is found in the crystal of the KI complex, including partial occupancy of phenylalanine at position 11. Only three sites in each of the two crystallographically independent channels are partially occupied by potassium cations instead of the commonly observed seven sites. The sum of the partial occupancies of K(+) (1.10 per two dimers) is consistent with the sum of the iodide occupancies (1.095 over eight sites), which is also confirmed by the anomalous signal of the iodide. There was a significant asymmetry of the distribution and occupancies of cations in the crystallographically independent gramicidin channels, in contrast to the distribution found in the rubidium chloride complex with gD.
Organizational Affiliation:
Institute of General and Ecological Chemistry, University of Technology, Łódź, Poland.