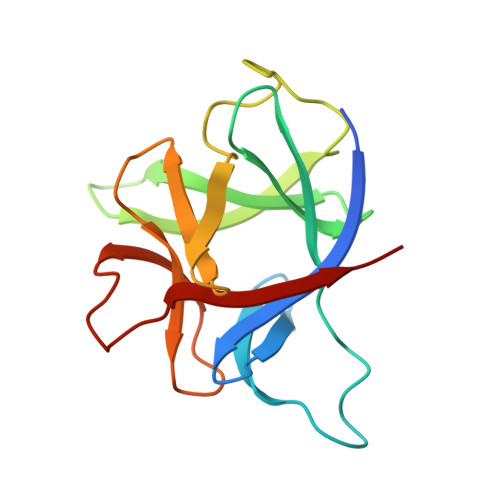
Initial crystallographic analysis of a recombinant human interleukin-1 receptor antagonist protein.
Clancy, L.L., Finzel, B.C., Yem, A.W., Deibel, M.R., Strakalaitis, N.A., Brunner, D.P., Sweet, R.M., Einspahr, H.M.(1994) Acta Crystallogr D Biol Crystallogr 50: 197-201
- PubMed: 15299459
- DOI: https://doi.org/10.1107/S0907444993009394
- Primary Citation of Related Structures:
2IRT - PubMed Abstract:
We report the crystallization of samples of a recombinant preparation of human interleukin-1 receptor antagonist protein (IRAP) and solution of the crystal structure by isomorphous replacement methods. Crystals were obtained by the hanging-drop vapor-diffusion method at 277 K from solutions of PEG 4000 containing sodium chloride, dithiothreitol and PIPES [sodium piperazione-N,N'-bis(2-ethanesulfonate)] buffer at pH 7.0. Crystals appear within about a week and grow as truncated tetragonal bipyramids to 0.3-0.6 mm on an edge. X-ray diffraction data from these crystals specify space group P4(3)2(1)2 and unit-cell dimensions of a = b = 72.35(26), c = 114.7(8) A and Z = 16 (two molecules per asymmetric unit). Fresh crystals diffract to about 2.3 A resolution. The search for heavy-atom derivatives has produced two, potassium gold cyanide and trimethyl lead chloride, as same-site, single-site derivatives. Inspection of an electron-density map at 4 A resolution calculated with these derivatives confirms that the IRAP molecule is a member of the interleukin-1 structural family.
Organizational Affiliation:
The Upjohn Company, Kalamazoo, Michigan 49001, USA.