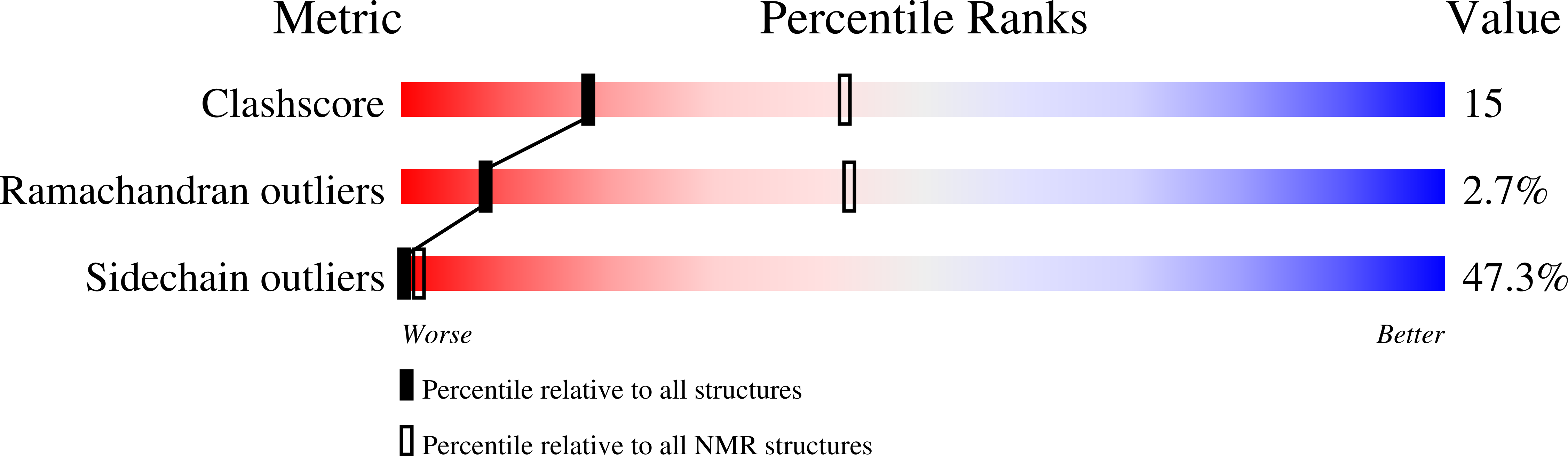Insights into transcription enhancer factor 1 (TEF-1) activity from the solution structure of the TEA domain.
Anbanandam, A., Albarado, D.C., Nguyen, C.T., Halder, G., Gao, X., Veeraraghavan, S.(2006) Proc Natl Acad Sci U S A 103: 17225-17230
- PubMed: 17085591
- DOI: https://doi.org/10.1073/pnas.0607171103
- Primary Citation of Related Structures:
2HZD - PubMed Abstract:
Transcription enhancer factor 1 is essential for cardiac, skeletal, and smooth muscle development and uses its N-terminal TEA domain (TEAD) to bind M-CAT elements. Here, we present the first structure of TEAD and show that it is a three-helix bundle with a homeodomain fold. Structural data reveal how TEAD binds DNA. Using structure-function correlations, we find that the L1 loop is essential for cooperative loading of TEAD molecules on to tandemly duplicated M-CAT sites. Furthermore, using a microarray chip-based assay, we establish that known binding sites of the full-length protein are only a subset of DNA elements recognized by TEAD. Our results provide a model for understanding the regulation of genome-wide gene expression during development by TEA/ATTS family of transcription factors.
Organizational Affiliation:
Department of Biochemistry & Molecular Biology, University of Texas Medical School, Houston, TX 77030, USA.