Ligand-induced asymmetry in histidine sensor kinase complex regulates quorum sensing.
Neiditch, M.B., Federle, M.J., Pompeani, A.J., Kelly, R.C., Swem, D.L., Jeffrey, P.D., Bassler, B.L., Hughson, F.M.(2006) Cell 126: 1095-1108
- PubMed: 16990134
- DOI: https://doi.org/10.1016/j.cell.2006.07.032
- Primary Citation of Related Structures:
2HJ9, 2HJE - PubMed Abstract:
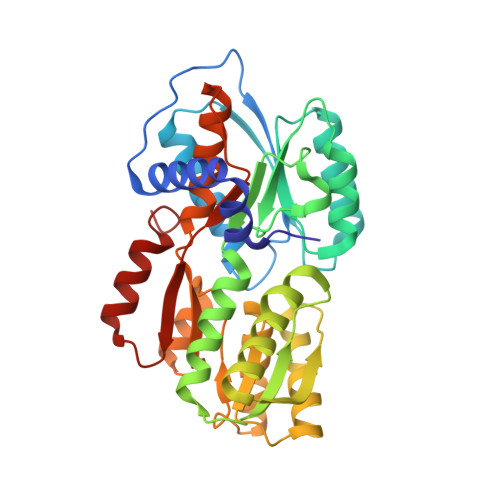
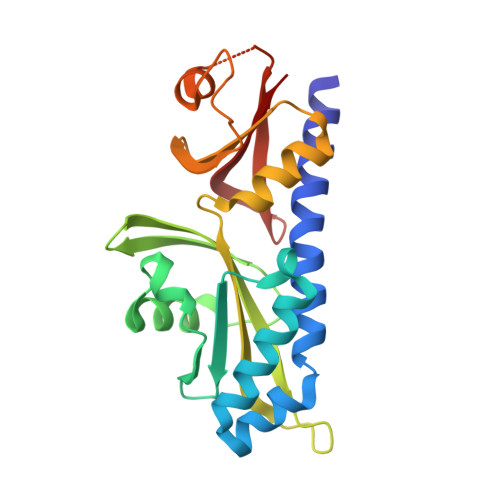
Bacteria sense their environment using receptors of the histidine sensor kinase family, but how kinase activity is regulated by ligand binding is not well understood. Autoinducer-2 (AI-2), a secreted signaling molecule originally identified in studies of the marine bacterium Vibrio harveyi, regulates quorum-sensing responses and allows communication between different bacterial species. AI-2 signal transduction in V. harveyi requires the integral membrane receptor LuxPQ, comprised of periplasmic binding protein (LuxP) and histidine sensor kinase (LuxQ) subunits. Combined X-ray crystallographic and functional studies show that AI-2 binding causes a major conformational change within LuxP, which in turn stabilizes a quaternary arrangement in which two LuxPQ monomers are asymmetrically associated. We propose that formation of this asymmetric quaternary structure is responsible for repressing the kinase activity of both LuxQ subunits and triggering the transition of V. harveyi into quorum-sensing mode.
Organizational Affiliation:
Department of Molecular Biology, Princeton University, Princeton, NJ 08544, USA.