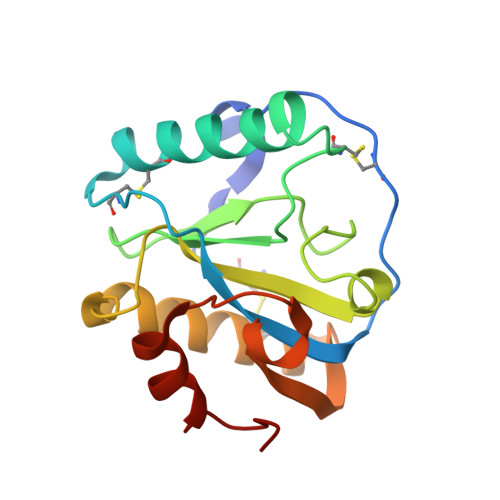
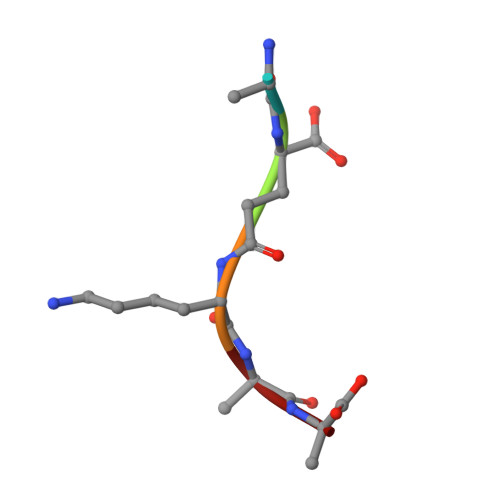
Structural insights into the bactericidal mechanism of human peptidoglycan recognition proteins
Cho, S., Wang, Q., Swaminathan, C.P., Hesek, D., Lee, M., Boons, G.J., Mobashery, S., Mariuzza, R.A.(2007) Proc Natl Acad Sci U S A 104: 8761-8766
- PubMed: 17502600
- DOI: https://doi.org/10.1073/pnas.0701453104
- Primary Citation of Related Structures:
2EAV, 2EAX - PubMed Abstract:
Peptidoglycan recognition proteins (PGRPs) are highly conserved pattern-recognition molecules of the innate immune system that bind bacterial peptidoglycans (PGNs), which are polymers of alternating N-acetylglucosamine (NAG) and N-acetylmuramic acid (NAM) cross-linked by short peptide stems. Human PRGPs are bactericidal against pathogenic and nonpathogenic Gram-positive bacteria, but not normal flora bacteria. Like certain glycopeptide antibiotics (e.g., vancomycin), PGRPs kill bacteria by directly interacting with their cell wall PGN, thereby interfering with PGN maturation. To better understand the bactericidal mechanism of PGRPs, we determined the crystal structure of the C-terminal PGN-binding domain of human PGRP-I beta in complex with NAG-NAM-L-Ala-gamma-D-Glu-L-Lys-D-Ala-D-Ala, a synthetic glycopeptide comprising a complete PGN repeat. This structure, in conjunction with the previously reported NMR structure of a dimeric PGN fragment, permitted identification of major conformational differences between free and PGRP-bound PGN with respect to the relative orientation of saccharide and peptide moieties. These differences provided structural insights into the bactericidal mechanism of human PGRPs. On the basis of molecular modeling, we propose that these proteins disrupt cell wall maturation not only by sterically encumbering access of biosynthetic enzymes to the nascent PGN chains, but also by locking PGN into a conformation that prevents formation of cross-links between peptide stems in the growing cell wall.
Organizational Affiliation:
Center for Advanced Research in Biotechnology, W. M. Keck Laboratory for Structural Biology, University of Maryland Biotechnology Institute, 9600 Gudelsky Drive, Rockville, MD 20850, USA.