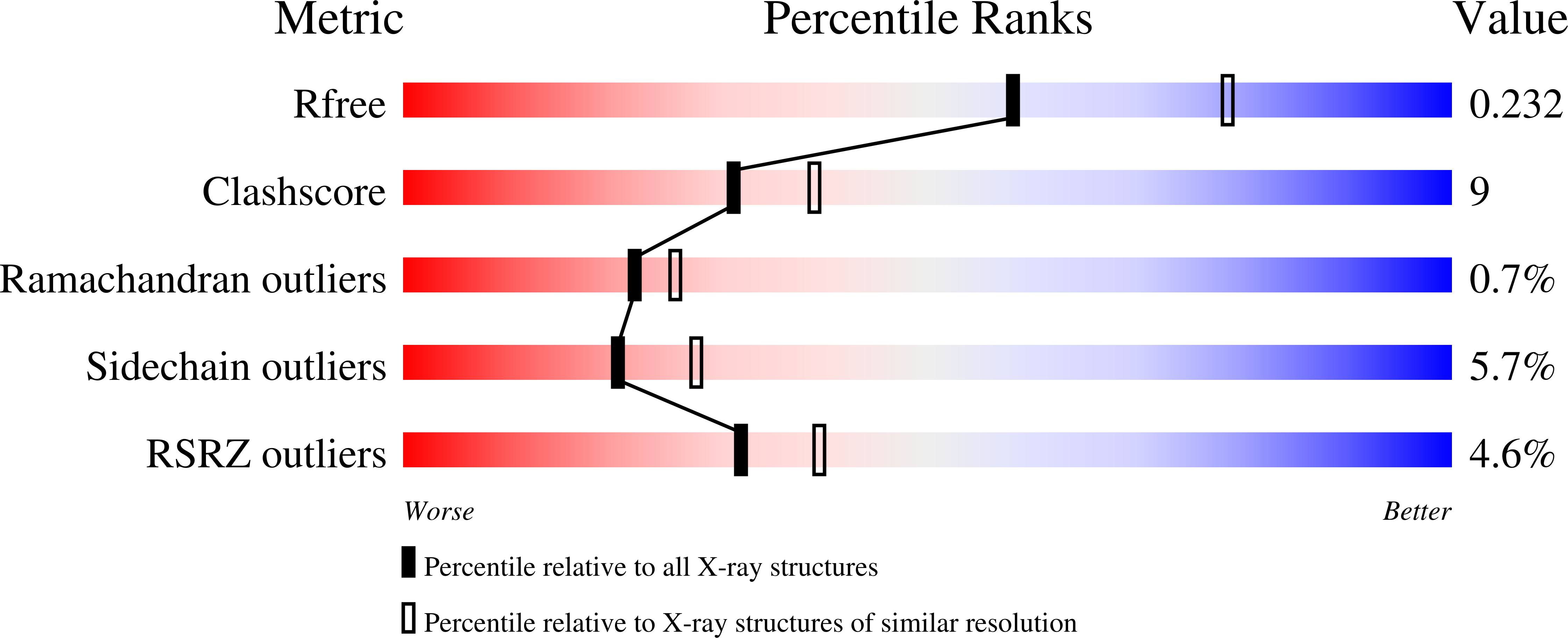
Structure of a lectin from Canavalia gladiata seeds: new structural insights for old molecules
Delatorre, P., Rocha, B.A.M., Souza, E.P., Oliveira, T.M., Bezerra, G.A., Moreno, F.B.M.B., Freitas, B.T., Santi-Gadelha, T., Sampaio, A.H., Azevedo Jr., W.F., Cavada, B.S.(2007) BMC Struct Biol 7: 52-52
- PubMed: 17683532
- DOI: https://doi.org/10.1186/1472-6807-7-52
- Primary Citation of Related Structures:
1WUV, 2D7F - PubMed Abstract:
Lectins are mainly described as simple carbohydrate-binding proteins. Previous studies have tried to identify other binding sites, which possible recognize plant hormones, secondary metabolites, and isolated amino acid residues. We report the crystal structure of a lectin isolated from Canavalia gladiata seeds (CGL), describing a new binding pocket, which may be related to pathogen resistance activity in ConA-like lectins; a site where a non-protein amino-acid, alpha-aminobutyric acid (Abu), is bound. The overall structure of native CGL and complexed with alpha-methyl-mannoside and Abu have been refined at 2.3 A and 2.31 A resolution, respectively. Analysis of the electron density maps of the CGL structure shows clearly the presence of Abu, which was confirmed by mass spectrometry. The presence of Abu in a plant lectin structure strongly indicates the ability of lectins on carrying secondary metabolites. Comparison of the amino acids composing the site with other legume lectins revealed that this site is conserved, providing an evidence of the biological relevance of this site. This new action of lectins strengthens their role in defense mechanisms in plants.
Organizational Affiliation:
Departamento de Bioquímica e Biologia Molecular, Universidade Federal do Ceará, Ceará, Brazil. plinio@urca.br