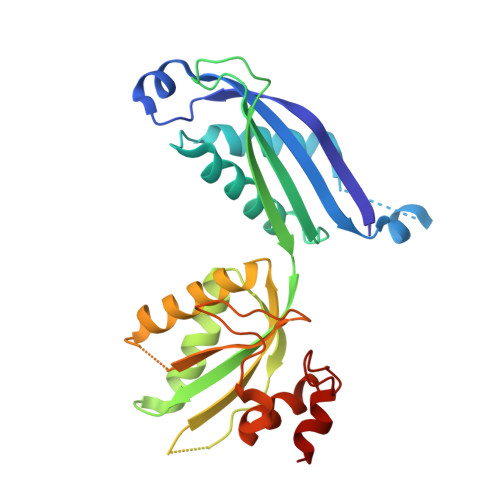
Crystal Structure of the Bifunctional Dihydroneopterin Aldolase/6-Hydroxymethyl-7,8-Dihydropterin Pyrophosphokinase from Streptococcus Pneumoniae.
Garcon, A., Levy, C., Derrick, J.P.(2006) J Mol Biol 360: 644
- PubMed: 16781731
- DOI: https://doi.org/10.1016/j.jmb.2006.05.038
- Primary Citation of Related Structures:
2CG8 - PubMed Abstract:
The enzymes dihydroneopterin aldolase (DHNA) and 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase (HPPK) catalyse two consecutive steps in the biosynthesis of folic acid. Neither of these enzymes has a counterpart in mammals, and they have therefore been suggested as ideal targets for antimicrobial drugs. Some of the enzymes within the folate pathway can occur as bi- or trifunctional complexes in bacteria and parasites, but the way in which bifunctional DHNA-HPPK enzymes are assembled is unclear. Here, we report the determination of the structure at 2.9 A resolution of the DHNA-HPPK (SulD) bifunctional enzyme complex from the respiratory pathogen Streptococcus pneumoniae. In the crystal, DHNA is assembled as a core octamer, with 422 point group symmetry, although the enzyme is active as a tetramer in solution. Individual HPPK monomers are arranged at the ends of the DHNA octamer, making relatively few contacts with the DHNA domain, but more extensive interactions with adjacent HPPK domains. As a result, the structure forms an elongated cylinder, with the HPPK domains forming two tetramers at each end. The active sites of both enzymes face outward, and there is no clear channel between them that could be used for channelling substrates. The HPPK-HPPK interface accounts for about one-third of the total area between adjacent monomers in SulD, and has levels of surface complementarity comparable to that of the DHNA-DHNA interfaces. There is no "linker" polypeptide between DHNA and HPPK, reducing the conformational flexibility of the HPPK domain relative to the DHNA domain. The implications for the organisation of bi- and trifunctional enzyme complexes within the folate biosynthesis pathway are discussed.
Organizational Affiliation:
Faculty of Life Sciences, The University of Manchester, Manchester Interdisciplinary Biocentre, 131 Princess Street, Manchester M1 7ND, UK.