Determination of the nuclear-magnetic-resonance solution structure of cardiotoxin CTX IIb from Naja mossambica mossambica.
O'Connell, J.F., Bougis, P.E., Wuthrich, K.(1993) Eur J Biochem 213: 891-900
- PubMed: 8504828
- DOI: https://doi.org/10.1111/j.1432-1033.1993.tb17833.x
- Primary Citation of Related Structures:
2CCX - PubMed Abstract:
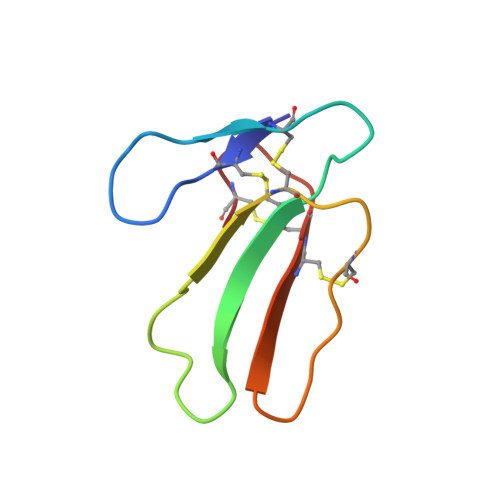
The NMR structure of cardiotoxin CTX IIb from Naja mossambica mossambica in aqueous solution was determined from a total of 593 nuclear Overhauser enhancement distance constraints and 135 dihedral angle constraints, which were collected using two-dimensional homonuclear 1H-NMR experiments. Structure calculations were performed with the program DIANA, using the redundant dihedral angle constraints strategy for improved convergence, followed by restrained energy minimization with the program FANTOM and a modified version of the program AMBER. The CTX IIb structure is represented by a group of 20 conformers with an average root-mean-square deviation relative to the mean solution structure of 0.072 nm for the backbone atoms, and 0.116 nm for all heavy atoms. The molecular structure of CTX IIb is characterized by a three-stranded beta-sheet made up of residues 20-26, 32-39 and 48-54, and a two-stranded beta-sheet composed of residues 1-5 and 10-14. A cluster of four disulfide bonds, 3-21, 14-38, 42-53 and 54-59, form the core of the molecule and crosslink the individual polypeptide strands. The NMR structure is similar to the previously reported X-ray crystal structure of the cardiotoxin CTX VII4 from the same species. Differences between the two structures were noted in the tips of the two loops formed by residues 6-9 and 27-31, which connect the beta-strand 1-5 with 10-14, and 20-26 with 32-39, respectively. For these loops the NMR data also indicate significantly increased dynamic disorder in the solution structure. These observations are discussed with respect to earlier suggestions by others that these two loops are essential structural elements for function and specificity of a wide variety of homologous toxins.
Organizational Affiliation:
Institut für Molekularbiologie und Biophysik, Eidgenössische Technische Hochschule-Hönggerberg, Zürich, Switzerland.