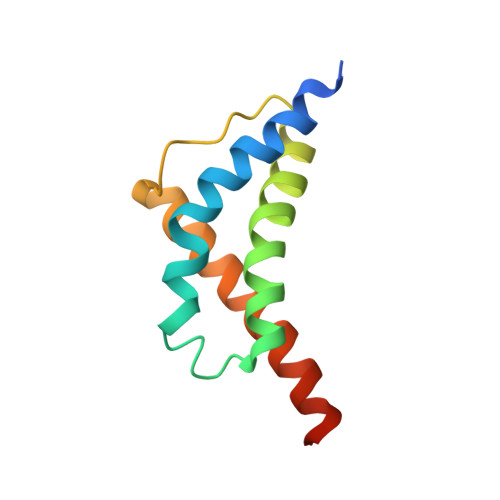
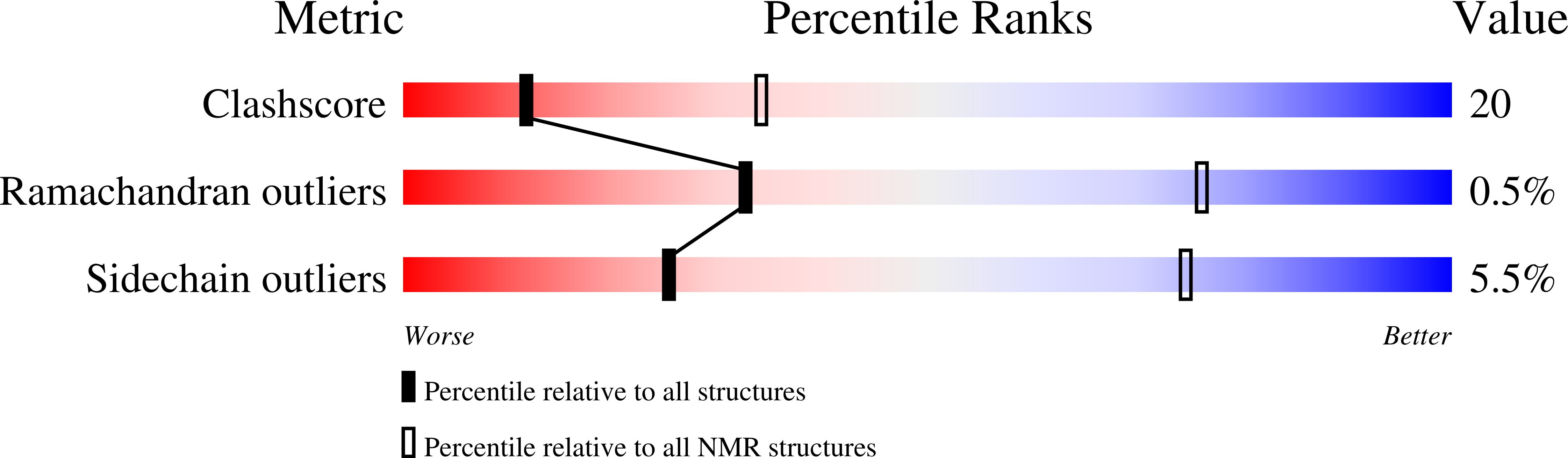Structure and carboxyl-terminal domain (CTD) binding of the Set2 SRI domain that couples histone H3 Lys36 methylation to transcription.
Vojnic, E., Simon, B., Strahl, B.D., Sattler, M., Cramer, P.(2006) J Biol Chem 281: 13-15
- PubMed: 16286474
- DOI: https://doi.org/10.1074/jbc.C500423200
- Primary Citation of Related Structures:
2C5Z - PubMed Abstract:
During mRNA elongation, the SRI domain of the histone H3 methyltransferase Set2 binds to the phosphorylated carboxyl-terminal domain (CTD) of RNA polymerase II. The solution structure of the yeast Set2 SRI domain reveals a novel CTD-binding fold consisting of a left-handed three-helix bundle. NMR titration shows that the SRI domain binds an Ser2/Ser5-phosphorylated CTD peptide comprising two heptapeptide repeats and three flanking NH2-terminal residues, whereas a single CTD repeat is insufficient for binding. Residues that show strong chemical shift perturbations upon CTD binding cluster in two regions. Both CTD tyrosine side chains contact the SRI domain. One of the tyrosines binds in the region with the strongest chemical shift perturbations, formed by the two NH2-terminal helices. Unexpectedly, the SRI domain fold resembles the structure of an RNA polymerase-interacting domain in bacterial sigma factors (domain sigma2 in sigma70).
Organizational Affiliation:
Gene Center, Department of Chemistry and Biochemistry, Ludwig-Maximilians-University of Munich, Feodor-Lynen-Strasse 25, 81377 Munich, Germany.