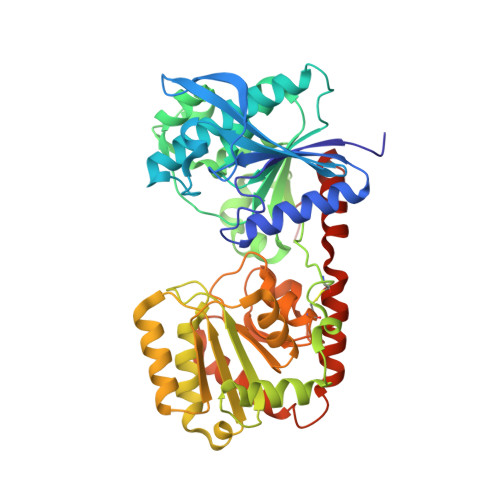
Crystal Structure of an Archaeal Glycogen Synthase: Insights Into Oligomerisation and Substrate Binding of Eukaryotic Glycogen Synthases.
Horcajada, C., Guinovart, J.J., Fita, I., Ferrer, J.C.(2006) J Biol Chem 281: 2923
- PubMed: 16319074
- DOI: https://doi.org/10.1074/jbc.M507394200
- Primary Citation of Related Structures:
2BFW, 2BIS - PubMed Abstract:
Glycogen and starch synthases are retaining glycosyltransferases that catalyze the transfer of glucosyl residues to the non-reducing end of a growing alpha-1,4-glucan chain, a central process of the carbon/energy metabolism present in almost all living organisms. The crystal structure of the glycogen synthase from Pyrococcus abyssi, the smallest known member of this family of enzymes, revealed that its subunits possess a fold common to other glycosyltransferases, a pair of beta/alpha/beta Rossmann fold-type domains with the catalytic site at their interface. Nevertheless, the archaeal enzyme presents an unprecedented homotrimeric molecular arrangement both in solution, as determined by analytical ultracentrifugation, and in the crystal. The C-domains are not involved in intersubunit interactions of the trimeric molecule, thus allowing for movements, likely required for catalysis, across the narrow hinge that connects the N- and C-domains. The radial disposition of the subunits confers on the molecule a distinct triangular shape, clearly visible with negative staining electron microscopy, in which the upper and lower faces present a sharp asymmetry. Comparison of bacterial and eukaryotic glycogen synthases, which use, respectively, ADP or UDP glucose as donor substrates, with the archaeal enzyme, which can utilize both molecules, allowed us to propose the residues that determine glucosyl donor specificity.
Organizational Affiliation:
Departament de Bioquímica i Biologia Molecular, Universitat de Barcelona, Spain.