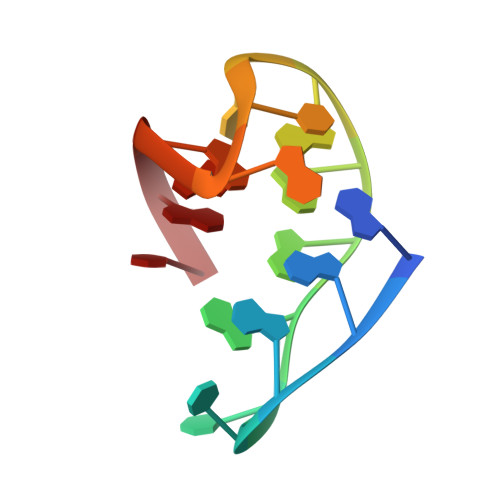
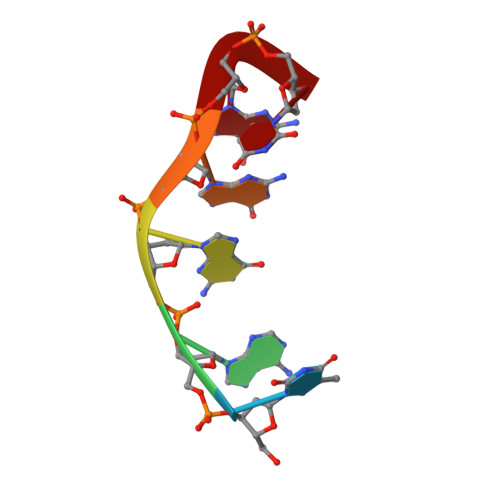
(3 + 1) Assembly of three human telomeric repeats into an asymmetric dimeric G-quadruplex
Zhang, N., Phan, A.T., Patel, D.J.(2005) J Am Chem Soc 127: 17277-17285
- PubMed: 16332077
- DOI: https://doi.org/10.1021/ja0543090
- Primary Citation of Related Structures:
2AQY - PubMed Abstract:
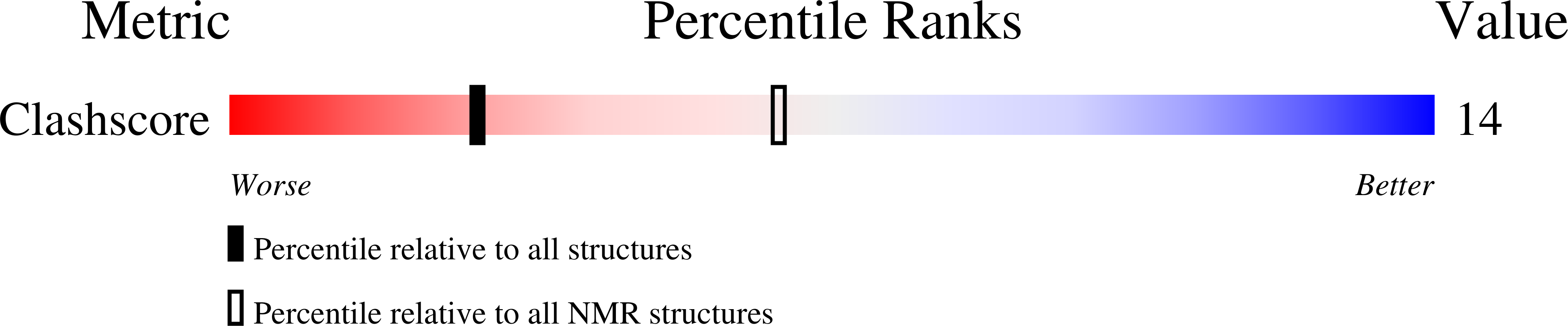
We present an NMR study on the structure of a DNA fragment of the human telomere containing three guanine-tracts, d(GGGTTAGGGTTAGGGT). This sequence forms in Na(+) solution a unique asymmetric dimeric quadruplex, in which the G-tetrad core involves all three G-tracts of one strand and only the last 3'-end G-tract of the other strand. We show that a three-repeat human telomeric sequence can also associate with a single-repeat human telomeric sequence into a structure with the same topology that we name (3 + 1) quadruplex assembly. In this G-quadruplex assembly, there are one syn.syn.syn.anti and two anti.anti.anti.syn G-tetrads, two edgewise loops, three G-tracts oriented in one direction and the fourth oriented in the opposite direction. We discuss the possible implications of the new folding topology for understanding the structure of telomeric DNA, including t-loop formation, and for targeting G-quadruplexes in the telomeres.
Organizational Affiliation:
Structural Biology Program, Memorial Sloan-Kettering Cancer Center, New York, New York 10021, USA.