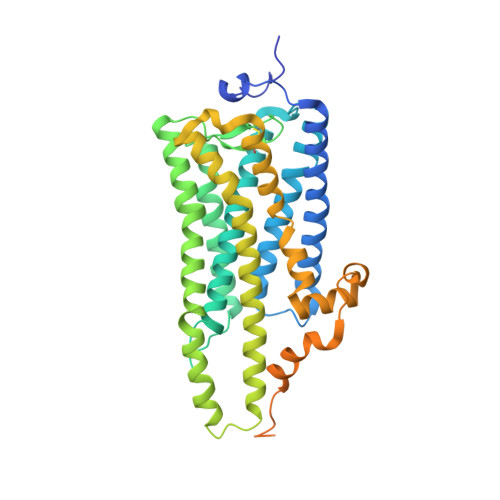
Crystal structure of squid rhodopsin.
Murakami, M., Kouyama, T.(2008) Nature 453: 363-367
- PubMed: 18480818
- DOI: https://doi.org/10.1038/nature06925
- Primary Citation of Related Structures:
2Z73 - PubMed Abstract:
Invertebrate phototransduction uses an inositol-1,4,5-trisphosphate signalling cascade in which photoactivated rhodopsin stimulates a G(q)-type G protein, that is, a class of G protein that stimulates membrane-bound phospholipase Cbeta. The same cascade is used by many G-protein-coupled receptors, indicating that invertebrate rhodopsin is a prototypical member. Here we report the crystal structure of squid (Todarodes pacificus) rhodopsin at 2.5 A resolution. Among seven transmembrane alpha-helices, helices V and VI extend into the cytoplasmic medium and, together with two cytoplasmic helices, they form a rigid protrusion from the membrane surface. This peculiar structure, which is not seen in bovine rhodopsin, seems to be crucial for the recognition of G(q)-type G proteins. The retinal Schiff base forms a hydrogen bond to Asn 87 or Tyr 111; it is far from the putative counterion Glu 180. In the crystal, a tight association is formed between the amino-terminal polypeptides of neighbouring monomers; this intermembrane dimerization may be responsible for the organization of hexagonally packed microvillar membranes in the photoreceptor rhabdom.
Organizational Affiliation:
Department of Physics, Graduate School of Science, Nagoya University, Nagoya 464-8602, Japan.