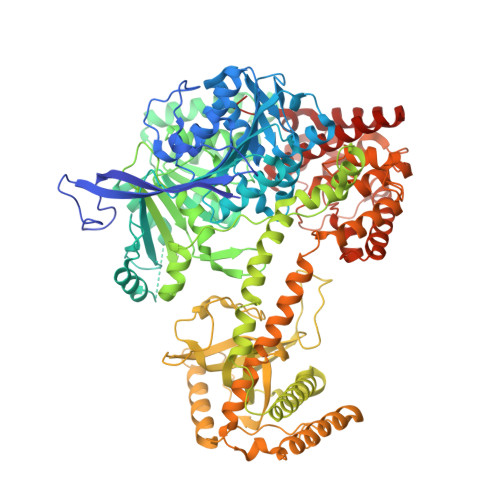
Structural Analysis Reveals the Characteristic Features of Mtr4, a Dexh Helicase Involved in Nuclear RNA Processing and Surveillance.
Weir, J.R., Bonneau, F., Hentschel, J., Conti, E.(2010) Proc Natl Acad Sci U S A 107: 12139
- PubMed: 20566885
- DOI: https://doi.org/10.1073/pnas.1004953107
- Primary Citation of Related Structures:
2XGJ - PubMed Abstract:
Mtr4 is a conserved RNA helicase that functions together with the nuclear exosome. It participates in the processing of structured RNAs, including the maturation of 5.8S ribosomal RNA (rRNA). It also interacts with the polyadenylating Trf4-Air2 heterodimer to form the so-called TRAMP (Trf4-Air2-Mtr4 Polyadenylation) complex. TRAMP is involved in exosome-mediated degradation of aberrant RNAs in nuclear surveillance pathways. We report the 2.9-A resolution crystal structure of Saccharomyces cerevisiae Mtr4 in complex with ADP and RNA. The structure shows a central ATPase core similar to that of other DExH helicases. Inserted in the DExH core is a region characteristic of Mtr4 orthologues that folds into an elongated stalk connected to a beta-barrel domain. This domain shows unexpected similarity to the KOW domain of L24, a ribosomal protein that binds 23S rRNA. We find that indeed the KOW domain of Mtr4 is able to bind in vitro transcribed tRNA(iMet), suggesting it might assist in presenting RNA substrates to the helicase core. The interaction of Mtr4 with Trf4-Air2 is mediated not by the stalk/KOW insertion but by the DExH core. We find that in the context of the TRAMP complex, the DExH core functions independently in vitro as an RNA helicase and a protein-binding platform. Mtr4 has thus evolved specific structural and surface features to perform its multiple functions.
Organizational Affiliation:
Department of Structural Cell Biology, Max-Planck-Institute of Biochemistry, Am Klopferspitz 18, D-82152 Martinsried, Germany.