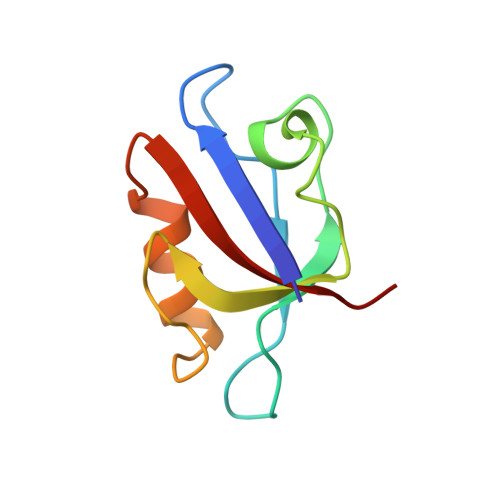
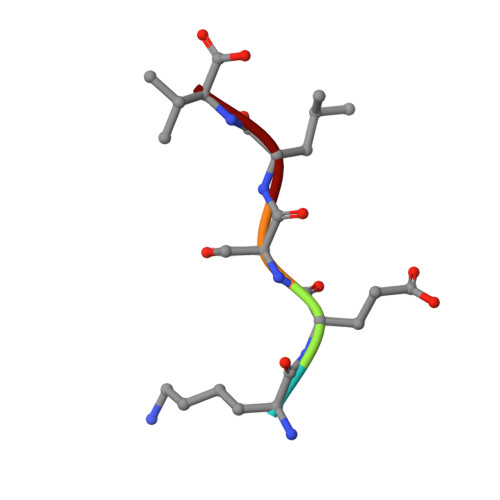
Specific interactions between the syntrophin PDZ domain and voltage-gated sodium channels.
Schultz, J., Hoffmuller, U., Krause, G., Ashurst, J., Macias, M.J., Schmieder, P., Schneider-Mergener, J., Oschkinat, H.(1998) Nat Struct Biol 5: 19-24
- PubMed: 9437424
- DOI: https://doi.org/10.1038/nsb0198-19
- Primary Citation of Related Structures:
2PDZ - PubMed Abstract:
Syntrophins are modular proteins belonging to the dystrophin associated glycoprotein complex and are thought to be involved in the regulation of the muscular system. Screening of peptide libraries revealed selectivity of the synotrophin PDZ domain toward the motif R/K/Q-E-S/T-X-V-COO- found to be highly conserved in the alpha-subunit C-terminus of vertebrate voltage gated sodium channels (VGSCs). The solution structure of the domain in complex with the peptide G-V-K-E-S-L-V shows specific interactions between the conserved residues in the peptide and syntrophin-characteristic residues in the domain. We propose that syntrophins localize VGSCs to the dystrophin network through its PDZ domain.
Organizational Affiliation:
Forschungsinstitut für Molekulare Pharmakologie, Berlin, Germany.