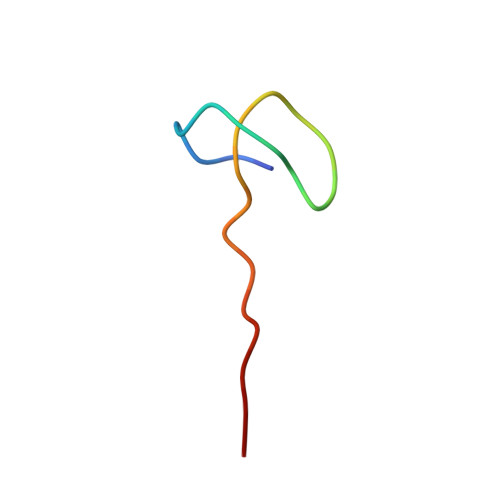
Elucidating the Specificity Determinants of the AtxE2 Lasso Peptide Isopeptidase.
Maksimov, M.O., Koos, J.D., Zong, C., Lisko, B., Link, A.J.(2015) J Biol Chem 290: 30806-30812
- PubMed: 26534965
- DOI: https://doi.org/10.1074/jbc.M115.694083
- Primary Citation of Related Structures:
2N6U, 2N6V - PubMed Abstract:
Lasso peptide isopeptidase is an enzyme that specifically hydrolyzes the isopeptide bond of lasso peptides, rendering these peptides linear. To carry out a detailed structure-activity analysis of the lasso peptide isopeptidase AtxE2 from Asticcacaulis excentricus, we solved NMR structures of its substrates astexin-2 and astexin-3. Using in vitro enzyme assays, we show that the C-terminal tail portion of these peptides is dispensable with regards to isopeptidase activity. A collection of astexin-2 and astexin-3 variants with alanine substitutions at each position within the ring and the loop was constructed, and we showed that all of these peptides except for one were cleaved by the isopeptidase. Thus, much like the lasso peptide biosynthetic enzymes, lasso peptide isopeptidase has broad substrate specificity. Quantitative analysis of the cleavage reactions indicated that alanine substitutions in loop positions of these peptides led to reduced cleavage, suggesting that the loop is serving as a recognition element for the isopeptidase.
Organizational Affiliation:
From the Departments of Chemical and Biological Engineering.