Structural and Functional Elucidation of Peptide Ts11 Shows Evidence of a Novel Subfamily of Scorpion Venom Toxins.
Cremonez, C.M., Maiti, M., Peigneur, S., Cassoli, J.S., Dutra, A.A., Waelkens, E., Lescrinier, E., Herdewijn, P., de Lima, M.E., Pimenta, A.M., Arantes, E.C., Tytgat, J.(2016) Toxins (basel) 8
- PubMed: 27706049
- DOI: https://doi.org/10.3390/toxins8100288
- Primary Citation of Related Structures:
2MSF - PubMed Abstract:
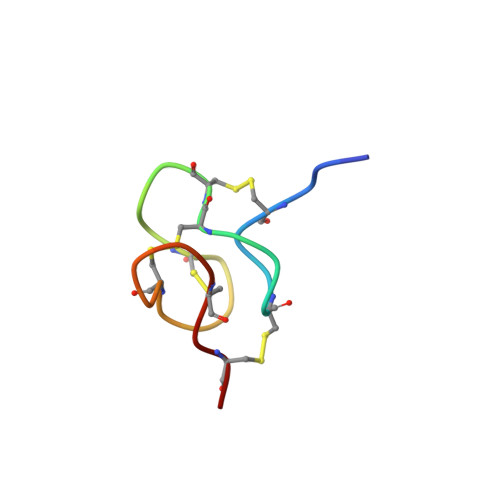
To date, several families of peptide toxins specifically interacting with ion channels in scorpion venom have been described. One of these families comprise peptide toxins (called KTxs), known to modulate potassium channels. Thus far, 202 KTxs have been reported, belonging to several subfamilies of KTxs (called α, β, γ, κ, δ, and λ-KTxs). Here we report on a previously described orphan toxin from Tityus serrulatus venom, named Ts11. We carried out an in-depth structure-function analysis combining 3D structure elucidation of Ts11 and electrophysiological characterization of the toxin. The Ts11 structure is highlighted by an Inhibitor Cystine Knot (ICK) type scaffold, completely devoid of the classical secondary structure elements (α-helix and/or β-strand). This has, to the best of our knowledge, never been described before for scorpion toxins and therefore represents a novel, 6th type of structural fold for these scorpion peptides. On the basis of their preferred interaction with voltage-gated K channels, as compared to all the other targets tested, it can be postulated that Ts11 is the first member of a new subfamily, designated as ε-KTx.
Organizational Affiliation:
Laboratório de Toxinas Animais, Departamento de Física e Química, Faculdade de Ciências Farmacêuticas de Ribeirão Preto, Universidade de São Paulo (USP), Ribeirão Preto 14040-903, São Paulo, Brasil. carolmarroni@yahoo.com.br.