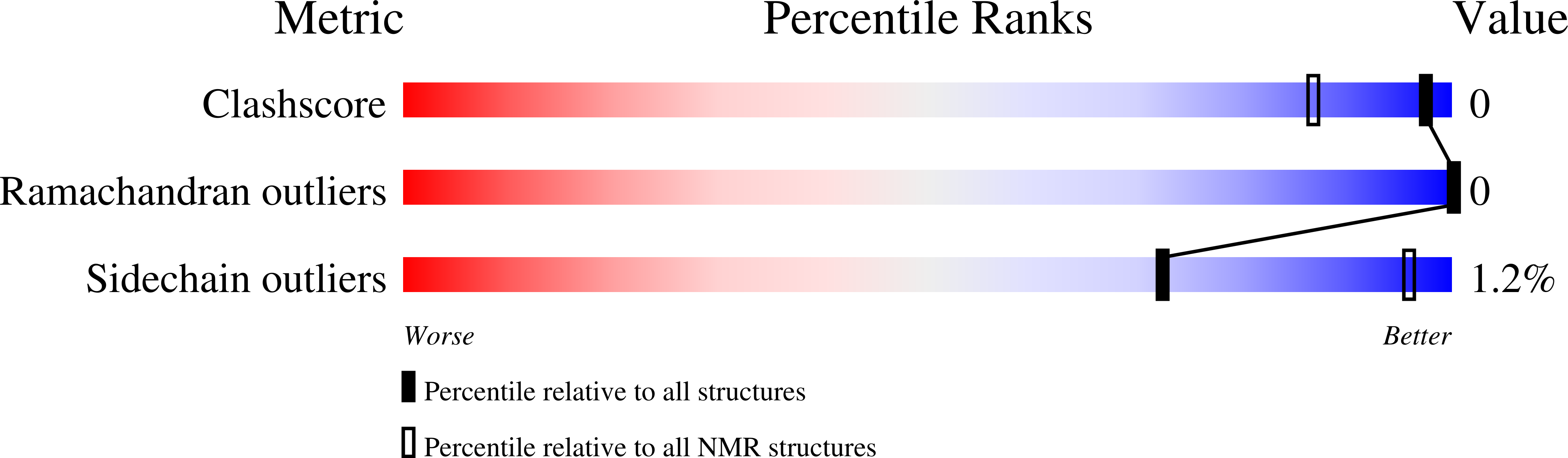Anticancer and toxic properties of cyclotides are dependent on phosphatidylethanolamine phospholipid targeting.
Henriques, S.T., Huang, Y.H., Chaousis, S., Wang, C.K., Craik, D.J.(2014) Chembiochem 15: 1956-1965
- PubMed: 25099014
- DOI: https://doi.org/10.1002/cbic.201402144
- Primary Citation of Related Structures:
2MN1 - PubMed Abstract:
Cyclotides, ultrastable disulfide-rich cyclic peptides, can be engineered to bind and inhibit specific cancer targets. In addition, some cyclotides are toxic to cancer cells, though not much is known about their mechanisms of action. Here we delineated the potential mode of action of cyclotides towards cancer cells. A novel set of analogues of kalata B1 (the prototypic cyclotide) and kalata B2 and cycloviolacin O2 were examined for their membrane-binding affinity and selectivity towards cancer cells. By using solution-state NMR, surface plasmon resonance, flow cytometry and bioassays we show that cyclotides are toxic against cancer and non-cancerous cells and their toxicity correlates with their ability to target and disrupt lipid bilayers that contain phosphatidylethanolamine phospholipids. Our results suggest that the potential of cyclotides as anticancer therapeutics might best be realised by combining their amenability to epitope engineering with their ability to bind cancer cell membranes.
Organizational Affiliation:
The University of Queensland, Institute for Molecular Bioscience, Carmody Road, St. Lucia, Brisbane, QLD 4072 (Australia). s.henriques@uq.edu.au.