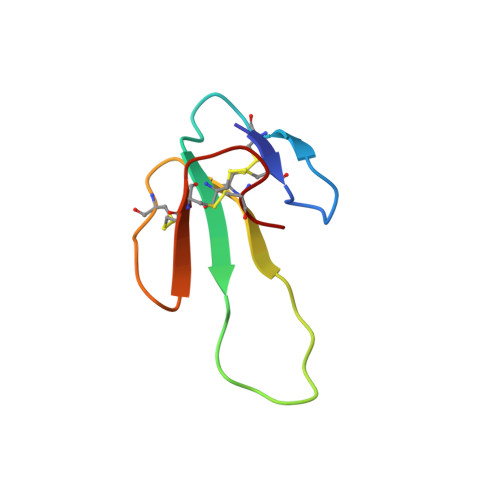
Chemical synthesis, 3D structure, and ASIC binding site of the toxin mambalgin-2.
Schroeder, C.I., Rash, L.D., Vila-Farres, X., Rosengren, K.J., Mobli, M., King, G.F., Alewood, P.F., Craik, D.J., Durek, T.(2014) Angew Chem Int Ed Engl 53: 1017-1020
- PubMed: 24323786
- DOI: https://doi.org/10.1002/anie.201308898
- Primary Citation of Related Structures:
2MFA - PubMed Abstract:
Mambalgins are a novel class of snake venom components that exert potent analgesic effects mediated through the inhibition of acid-sensing ion channels (ASICs). The 57-residue polypeptide mambalgin-2 (Ma-2) was synthesized by using a combination of solid-phase peptide synthesis and native chemical ligation. The structure of the synthetic toxin, determined using homonuclear NMR, revealed an unusual three-finger toxin fold reminiscent of functionally unrelated snake toxins. Electrophysiological analysis of Ma-2 on wild-type and mutant ASIC1a receptors allowed us to identify α-helix 5, which borders on the functionally critical acidic pocket of the channel, as a major part of the Ma-2 binding site. This region is also crucial for the interaction of ASIC1a with the spider toxin PcTx1, thus suggesting that the binding sites for these toxins substantially overlap. This work lays the foundation for structure-activity relationship (SAR) studies and further development of this promising analgesic peptide.
Organizational Affiliation:
Division of Chemistry and Structural Biology, Institute for Molecular Bioscience, The University of Queensland, Brisbane, Queensland (Australia).