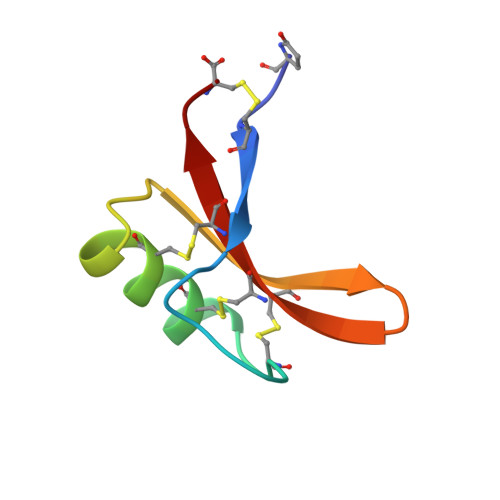
The nuclear magnetic resonance solution structure of the synthetic AhPDF1.1b plant defensin evidences the structural feature within the gamma-motif.
Meindre, F., Lelievre, D., Loth, K., Mith, O., Aucagne, V., Berthomieu, P., Marques, L., Delmas, A.F., Landon, C., Paquet, F.(2014) Biochemistry 53: 7745-7754
- PubMed: 25419866
- DOI: https://doi.org/10.1021/bi501285k
- Primary Citation of Related Structures:
2M8B - PubMed Abstract:
Plant defensins (PDF) are cysteine-rich peptides that are major actors in the innate immunity in plants. Besides their antifungal activity, some PDF such as Arabidopsis halleri PDF1.1b confer zinc tolerance in plants. Here we present (i) an efficient protocol for the production of AhPDF1.1b by solid-phase peptide synthesis followed by controlled oxidative folding to obtain the highly pure native form of the defensin and (ii) the three-dimensional (3D) nuclear magnetic resonance structure of AhPDF1.1b, the first 3D structure of plant defensin obtained with a synthetic peptide. Its fold is organized around the typical cysteine-stabilized α-helix β-sheet motif and contains the γ-core motif involved in the antifungal activity of all plant defensins. On the basis of our structural analysis of AhPDF1 defensins combined with previous biological data for antifungal and zinc tolerance activities, we established the essential role of cis-Pro41 within the γ-core. In fact, the four consecutive residues (Val39-Phe40-Pro41-Ala42) are strictly conserved for plant defensins able to tolerate zinc. We hypothesized that structural and/or dynamic features of this sequence are related to the ability of the defensin to chelate zinc.
Organizational Affiliation:
Centre de Biophysique Moléculaire, CNRS UPR4301 , Rue Charles Sadron, 45071 Orléans Cedex 2, France.