Structural similarity between defense peptide from wheat and scorpion neurotoxin permits rational functional design
Berkut, A.A., Usmanova, D.R., Peigneur, S., Oparin, P.B., Mineev, K.S., Odintsova, T.I., Tytgat, J., Arseniev, A.S., Grishin, E.V., Vassilevski, A.A.(2014) J Biol Chem 289: 14331-14340
- PubMed: 24671422
- DOI: https://doi.org/10.1074/jbc.M113.530477
- Primary Citation of Related Structures:
2M6A - PubMed Abstract:
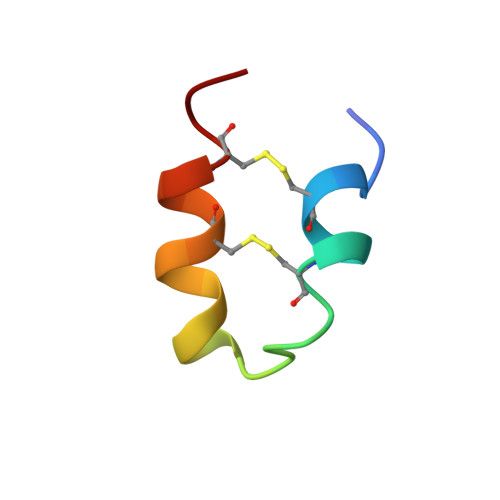
In this study, we present the spatial structure of the wheat antimicrobial peptide (AMP) Tk-AMP-X2 studied using NMR spectroscopy. This peptide was found to adopt a disulfide-stabilized α-helical hairpin fold and therefore belongs to the α-hairpinin family of plant defense peptides. Based on Tk-AMP-X2 structural similarity to cone snail and scorpion potassium channel blockers, a mutant molecule, Tk-hefu, was engineered by incorporating the functionally important residues from κ-hefutoxin 1 onto the Tk-AMP-X2 scaffold. The designed peptide contained the so-called essential dyad of amino acid residues significant for channel-blocking activity. Electrophysiological studies showed that although the parent peptide Tk-AMP-X2 did not present any activity against potassium channels, Tk-hefu blocked Kv1.3 channels with similar potency (IC50 ∼ 35 μm) to κ-hefutoxin 1 (IC50 ∼ 40 μm). We conclude that α-hairpinins are attractive in their simplicity as structural templates, which may be used for functional engineering and drug design.
Organizational Affiliation:
From the M. M. Shemyakin and Yu. A. Ovchinnikov Institute of Bioorganic Chemistry, Russian Academy of Sciences, Moscow 117997, Russia, Moscow Institute of Physics and Technology (State University), Moscow 117303, Russia.