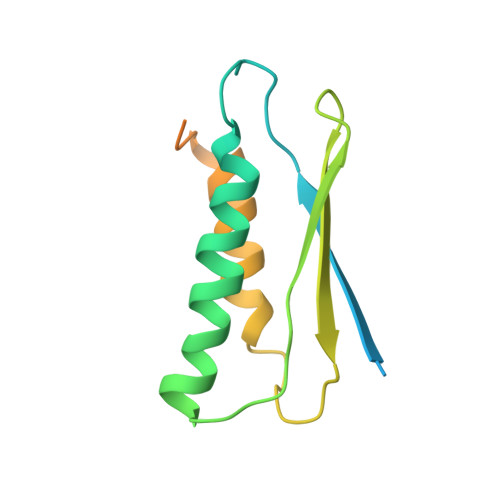
Solution NMR structure of a sheddase inhibitor prodomain from the malarial parasite Plasmodium falciparum.
He, Y., Chen, Y., Oganesyan, N., Ruan, B., O'Brochta, D., Bryan, P.N., Orban, J.(2012) Proteins 80: 2810-2817
- PubMed: 23011838
- DOI: https://doi.org/10.1002/prot.24187
- Primary Citation of Related Structures:
2LU1 - PubMed Abstract:
Plasmodium subtilisin 2 (Sub2) is a multidomain protein that plays an important role in malaria infection. Here, we describe the solution NMR structure of a conserved region of the inhibitory prodomain of Sub2 from Plasmodium falciparum, termed prosub2. Despite the absence of any detectable sequence homology, the protozoan prosub2 has structural similarity to bacterial and mammalian subtilisin-like prodomains. Comparison with the three-dimensional structures of these other prodomains suggests a likely binding interface with the catalytic domain of Sub2 and provides insights into the locations of primary and secondary processing sites in Plasmodium prodomains.
Organizational Affiliation:
Institute for Bioscience and Biotechnology Research, University of Maryland, Rockville, MD 20850, USA.