Structure and function of a unique pore-forming protein from a pathogenic acanthamoeba.
Michalek, M., Sonnichsen, F.D., Wechselberger, R., Dingley, A.J., Hung, C.W., Kopp, A., Wienk, H., Simanski, M., Herbst, R., Lorenzen, I., Marciano-Cabral, F., Gelhaus, C., Gutsmann, T., Tholey, A., Grotzinger, J., Leippe, M.(2013) Nat Chem Biol 9: 37-42
- PubMed: 23143413
- DOI: https://doi.org/10.1038/nchembio.1116
- Primary Citation of Related Structures:
2LRD, 2LRE - PubMed Abstract:
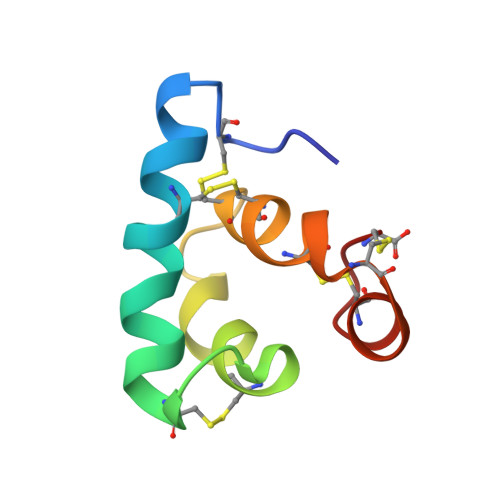
Human pathogens often produce soluble protein toxins that generate pores inside membranes, resulting in the death of target cells and tissue damage. In pathogenic amoebae, this has been exemplified with amoebapores of the enteric protozoan parasite Entamoeba histolytica. Here we characterize acanthaporin, to our knowledge the first pore-forming toxin to be described from acanthamoebae, which are free-living, bacteria-feeding, unicellular organisms that are opportunistic pathogens of increasing importance and cause severe and often fatal diseases. We isolated acanthaporin from extracts of virulent Acanthamoeba culbertsoni by tracking its pore-forming activity, molecularly cloned the gene of its precursor and recombinantly expressed the mature protein in bacteria. Acanthaporin was cytotoxic for human neuronal cells and exerted antimicrobial activity against a variety of bacterial strains by permeabilizing their membranes. The tertiary structures of acanthaporin's active monomeric form and inactive dimeric form, both solved by NMR spectroscopy, revealed a currently unknown protein fold and a pH-dependent trigger mechanism of activation.
Organizational Affiliation:
Zoological Institute, Zoophysiology, Christian-Albrechts Universität zu Kiel, Kiel, Germany.