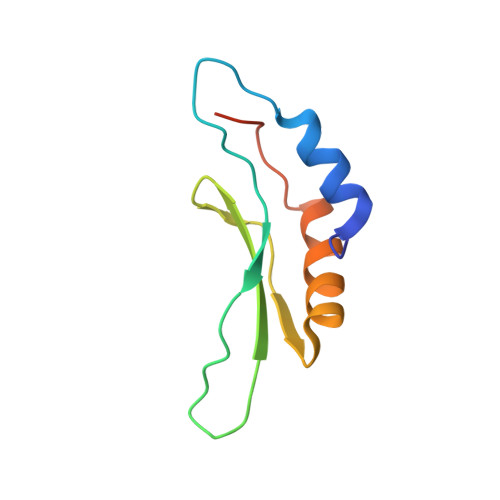
Solution structure of the Drosha double-stranded RNA-binding domain.
Mueller, G.A., Miller, M.T., Derose, E.F., Ghosh, M., London, R.E., Hall, T.M.(2010) Silence 1: 2-2
- PubMed: 20226070
- DOI: https://doi.org/10.1186/1758-907X-1-2
- Primary Citation of Related Structures:
2KHX - PubMed Abstract:
Drosha is a nuclear RNase III enzyme that initiates processing of regulatory microRNA. Together with partner protein DiGeorge syndrome critical region 8 (DGCR8), it forms the Microprocessor complex, which cleaves precursor transcripts called primary microRNA to produce hairpin precursor microRNA. In addition to two RNase III catalytic domains, Drosha contains a C-terminal double-stranded RNA-binding domain (dsRBD). To gain insight into the function of this domain, we determined the nuclear magnetic resonance (NMR) solution structure. We report here the solution structure of the dsRBD from Drosha (Drosha-dsRBD). The alphabetabetabetaalpha fold is similar to other dsRBD structures. A unique extended loop distinguishes this domain from other dsRBDs of known structure. Despite uncertainties about RNA-binding properties of the Drosha-dsRBD, its structure suggests it retains RNA-binding features. We propose that this domain may contribute to substrate recognition in the Drosha-DGCR8 Microprocessor complex.
Organizational Affiliation:
Laboratory of Structural Biology, National Institute of Environmental Health Sciences, National Institutes of Health, Research Triangle Park, NC, USA.