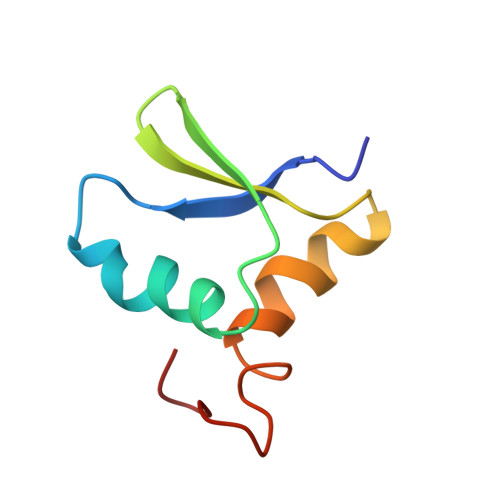
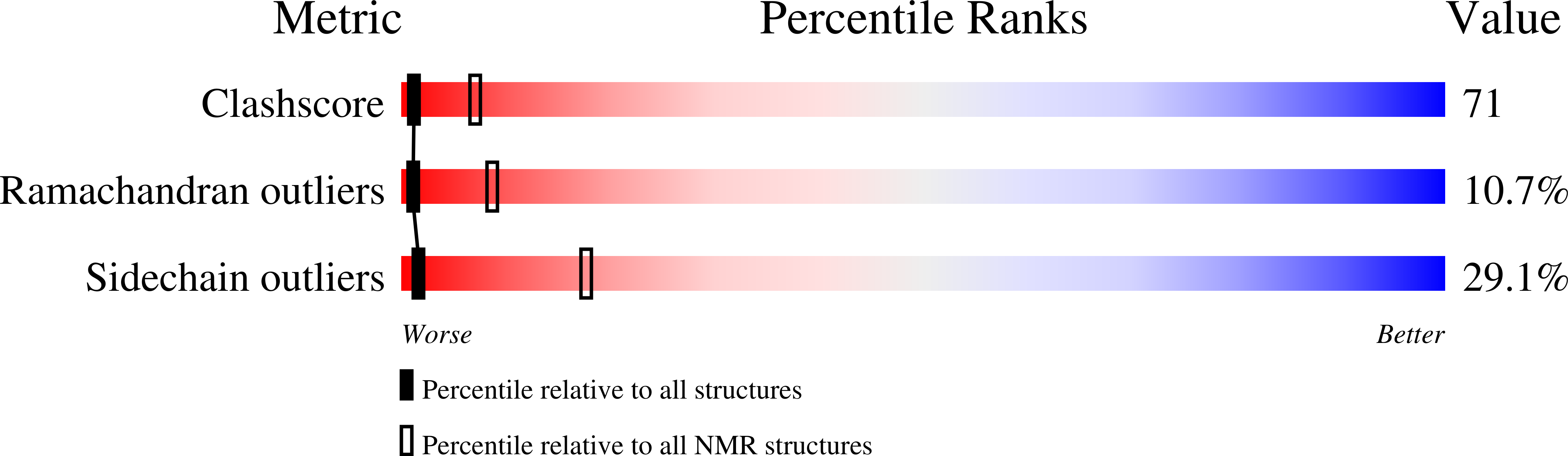Solution structure and dynamics of the N-terminal cytosolic domain of rhomboid intramembrane protease from Pseudomonas aeruginosa: insights into a functional role in intramembrane proteolysis.
Del Rio, A., Dutta, K., Chavez, J., Ubarretxena-Belandia, I., Ghose, R.(2007) J Mol Biol 365: 109-122
- PubMed: 17059825
- DOI: https://doi.org/10.1016/j.jmb.2006.09.047
- Primary Citation of Related Structures:
2GQC - PubMed Abstract:
Rhomboids are ubiquitous integral membrane proteases that release cellular signals from membrane-bound substrates through a general signal transduction mechanism known as regulated intramembrane proteolysis (RIP). We present the NMR structure of the cytosolic N-terminal domain (NRho) of P. aeruginosa Rhomboid. NRho consists of a novel alpha/beta fold and represents the first detailed structural insight into this class of intramembrane proteases. We find evidence that NRho is capable of strong and specific association with detergent micelles that mimic the membrane/water interface. Relaxation measurements on NRho reveal structural fluctuations on the microseconds-milliseconds timescale in regions including and contiguous to those implicated in membrane interaction. This structural plasticity may facilitate the ability of NRho to recognize and associate with membranes. We suggest that NRho plays a role in scissile peptide bond selectivity by optimally positioning the Rhomboid active site relative to the membrane plane.
Organizational Affiliation:
Department of Chemistry, The City College of New York, 138th Street & Convent Avenue, New York, NY 10031, USA.