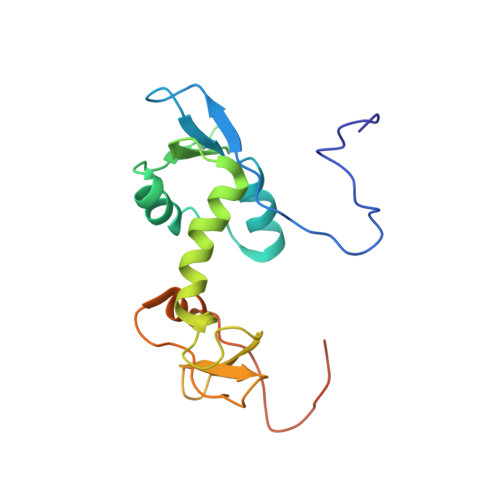
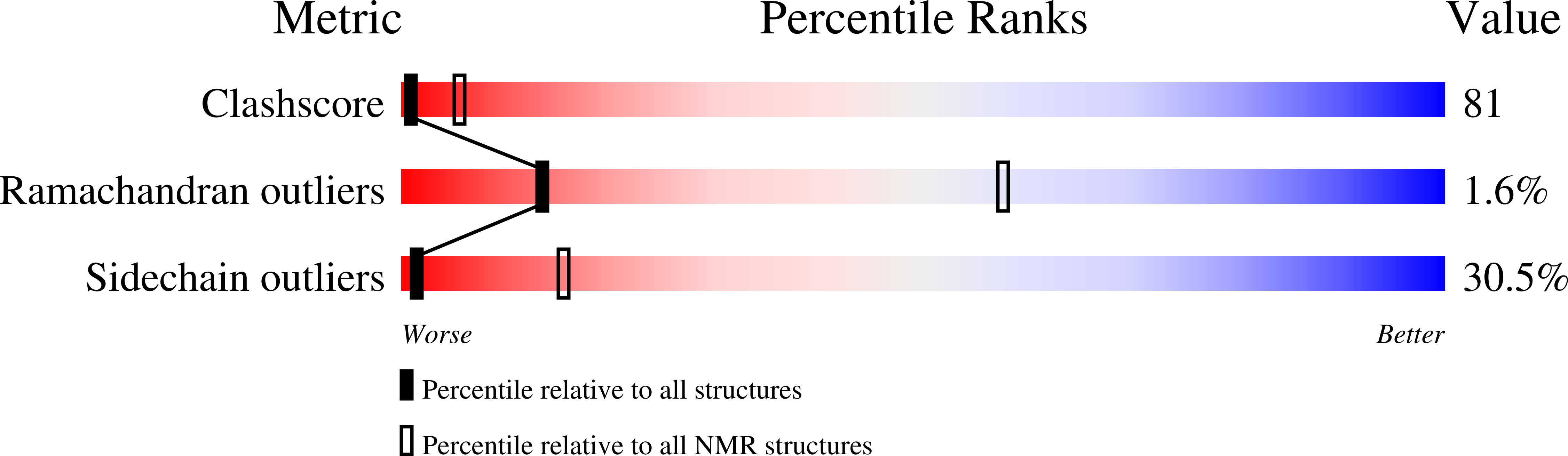Structure of the eukaryotic initiation factor (eIF) 5 reveals a fold common to several translation factors
Conte, M.R., Kelly, G., Babon, J., Sanfelice, D., Youell, J., Smerdon, S.J., Proud, C.G.(2006) Biochemistry 45: 4550-4558
- PubMed: 16584190
- DOI: https://doi.org/10.1021/bi052387u
- Primary Citation of Related Structures:
2G2K - PubMed Abstract:
Eukaryotic initiation factor 5 (eIF5) plays multiple roles in translation initiation. Its N-terminal domain functions as a GTPase-activator protein (GAP) for GTP bound to eIF2, while its C-terminal region nucleates the interactions between multiple translation factors, including eIF1, which acts to inhibit GTP hydrolysis or P(i) release, and the beta subunit of eIF2. These proteins and the events in which they participate are critical for the accurate recognition of the correct start codon during translation initiation. Here, we report the three-dimensional solution structure of the N-terminal domain of human eIF5, comprising two subdomains, both reminiscent of nucleic-acid-binding modules. The N-terminal subdomain contains the "arginine finger" motif that is essential for GAP function but which, unusually, resides in a partially disordered region of the molecule. This implies that a conformational reordering of this portion of eIF5 is likely to occur upon formation of a competent complex for GTP hydrolysis, following the appropriate activation signal. Interestingly, the N-terminal subdomain of eIF5 reveals an alpha/beta fold structurally similar to both the archaeal orthologue of the beta subunit of eIF2 and, unexpectedly, to eIF1. These results reveal a novel protein fold common to several factors involved in related steps of translation initiation. The implications of these observations are discussed in terms of the mechanism of translation initiation.
Organizational Affiliation:
Biophysics Laboratories, School of Biological Sciences, University of Portsmouth, Portsmouth PO1 2DT, United Kingdom.