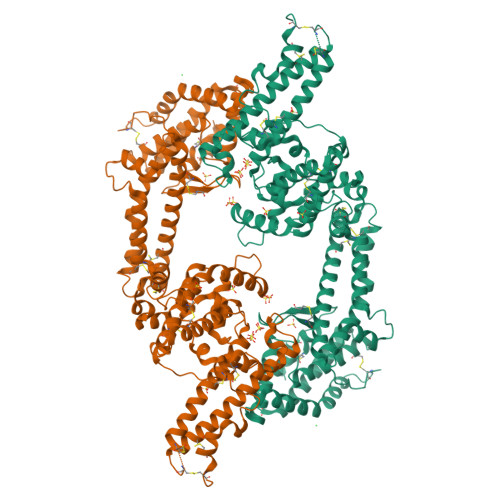
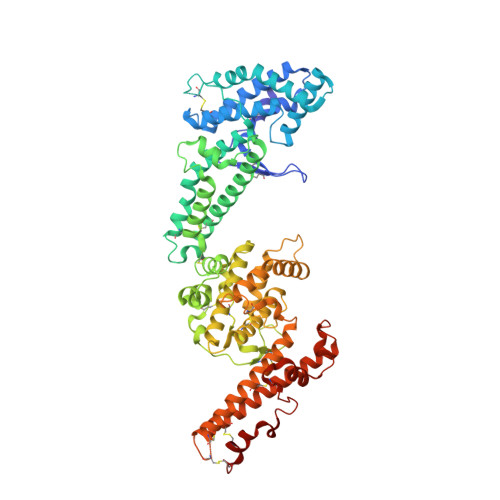
Structural Basis for the EBA-175 Erythrocyte Invasion Pathway of the Malaria Parasite Plasmodium falciparum.
Tolia, N.H., Enemark, E.J., Sim, B.K., Joshua-Tor, L.(2005) Cell 122: 183-193
- PubMed: 16051144
- DOI: https://doi.org/10.1016/j.cell.2005.05.033
- Primary Citation of Related Structures:
1ZRL, 1ZRO - PubMed Abstract:
Erythrocyte binding antigen 175 (EBA-175) is a P. falciparum protein that binds the major glycoprotein found on human erythrocytes, glycophorin A, during invasion. Here we present the crystal structure of the erythrocyte binding domain of EBA-175, RII, which has been established as a vaccine candidate. Binding sites for the heavily sialylated receptor glycophorin A are proposed based on a complex of RII with a glycan that contains the essential components required for binding. The dimeric organization of RII displays two prominent channels that contain four of the six observed glycan binding sites. Each monomer consists of two Duffy binding-like (DBL) domains (F1 and F2). F2 more prominently lines the channels and makes the majority of the glycan contacts, underscoring its role in cytoadherence and in antigenic variation in malaria. Our studies provide insight into the mechanism of erythrocyte invasion by the malaria parasite and aid in rational drug design and vaccines.
Organizational Affiliation:
Watson School of Biological Sciences, Cold Spring Harbor Laboratory, 1 Bungtown Road, Cold Spring Harbor, New York 11724, USA.