Solution Structure of Zinc- and Calcium-Bound Rat S100B as Determined by Nuclear Magnetic Resonance Spectroscopy
Wilder, P.T., Varney, K.M., Weiss, M.B., Gitti, R.K., Weber, D.J.(2005) Biochemistry 44: 5690-5702
- PubMed: 15823027
- DOI: https://doi.org/10.1021/bi0475830
- Primary Citation of Related Structures:
1XYD - PubMed Abstract:
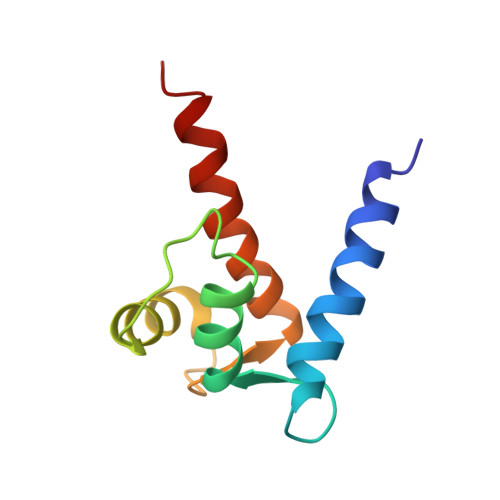
The EF-hand calcium-binding protein S100B also binds one zinc ion per subunit with a relatively high affinity (K(d) approximately 90 nM) [Wilder et al., (2003) Biochemistry 42, 13410-13421]. In this study, the structural characterization of zinc binding to calcium-loaded S100B was examined using high-resolution NMR techniques, including structural characterization of this complex in solution at atomic resolution. As with other S100 protein structures, the quaternary structure of Zn(2+)-Ca(2+)-bound S100B was found to be dimeric with helices H1, H1', H4, and H4' forming an X-type four-helix bundle at the dimer interface. NMR data together with mutational analyses are consistent with Zn(2+) coordination arising from His-15 and His-25 of one S100B subunit and from His-85 and Glu-89 of the other subunit. The addition of Zn(2+) was also found to extend helices H4 and H4' three to four residues similar to what was previously observed with the binding of target proteins to S100B. Furthermore, a kink in helix 4 was observed in Zn(2+)-Ca(2+)-bound S100B that is not in Ca(2+)-bound S100B. These structural changes upon Zn(2+)-binding could explain the 5-fold increase in affinity that Zn(2+)-Ca(2+)-bound S100B has for peptide targets such as the TRTK peptide versus Ca(2+)-bound S100B. There are also changes in the relative positioning of the two EF-hand calcium-binding domains and the respective helices comprising these EF-hands. Changes in conformation such as these could contribute to the order of magnitude higher affinity that S100B has for calcium in the presence of Zn(2+).
Organizational Affiliation:
Molecular and Cell Biology Program, University of Maryland School of Medicine, 108 North Greene Street, Baltimore, Maryland 21201, USA.