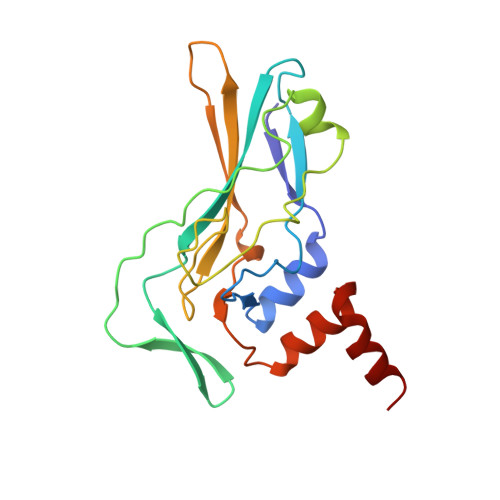
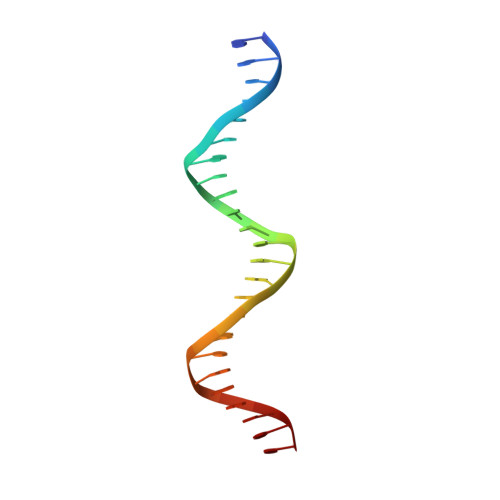
Crystallographic structure of the T domain-DNA complex of the Brachyury transcription factor.
Muller, C.W., Herrmann, B.G.(1997) Nature 389: 884-888
- PubMed: 9349824
- DOI: https://doi.org/10.1038/39929
- Primary Citation of Related Structures:
1XBR - PubMed Abstract:
The mouse Brachyury (T) gene is the prototype of a growing family of so-called T-box genes which encode transcriptional regulators and have been identified in a variety of invertebrates and vertebrates, including humans. Mutations in Brachyury and other T-box genes result in drastic embryonic phenotypes, indicating that T-box gene products are essential in tissue specification, morphogenesis and organogenesis. The T-box encodes a DNA-binding domain of about 180 amino-acid residues, the T domain. Here we report the X-ray structure of the T domain from Xenopus laevis in complex with a 24-nucleotide palindromic DNA duplex. We show that the protein is bound as a dimer, interacting with the major and the minor grooves of the DNA. A new type of specific DNA contact is seen, in which a carboxy-terminal helix is deeply embedded into an enlarged minor groove without bending the DNA. Hydrophobic interactions and an unusual main-chain carbonyl contact to a guanine account for sequence-specific recognition in the minor groove by this helix. Thus the structure of this T domain complex with DNA reveals a new way in which a protein can recognize DNA.
Organizational Affiliation:
European Molecular Biology Laboratory, Grenoble Outstation, France. mueller@embl-grenoble.fr