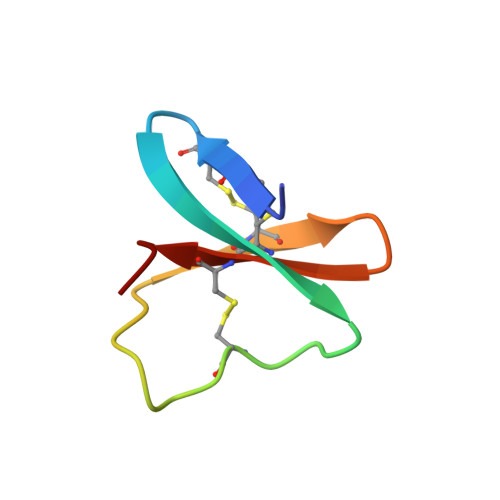
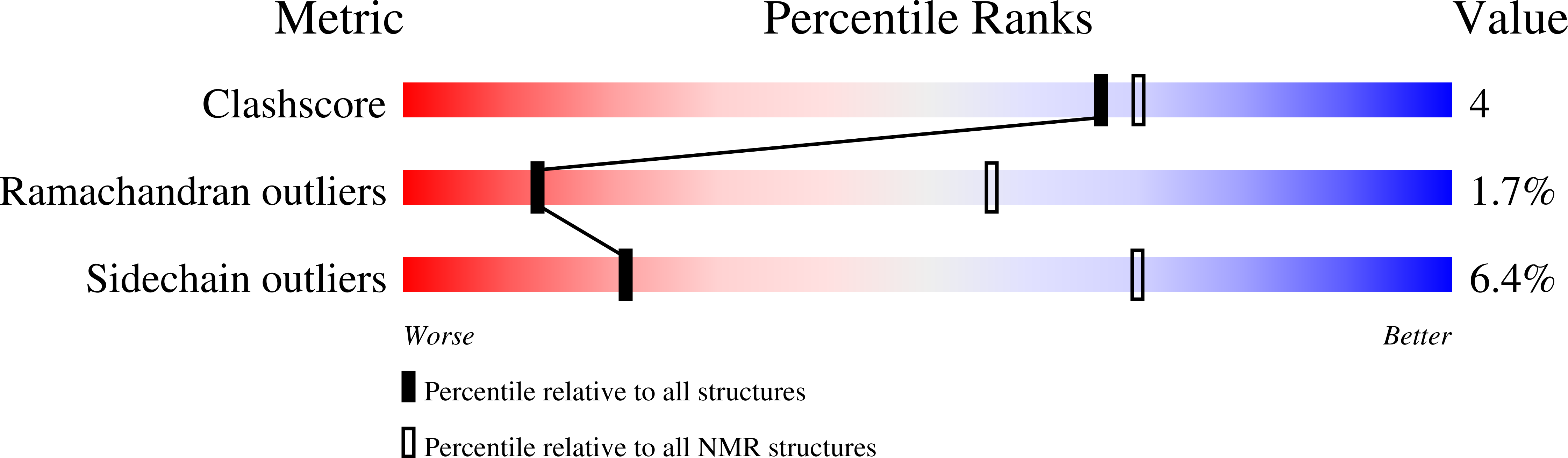Solution structure of APETx1 from the sea anemone Anthopleura elegantissima: A new fold for an HERG toxin
Chagot, B., Diochot, S., Pimentel, C., Lazdunski, M., Darbon, H.(2005) Proteins 59: 380-386
- PubMed: 15726634
- DOI: https://doi.org/10.1002/prot.20425
- Primary Citation of Related Structures:
1WQK - PubMed Abstract:
APETx1 is a 42-amino acid toxin purified from the venom of the sea anemone Anthopleura elegantissima. This cysteine-rich peptide possesses three disulfide bridges (C4-C37, C6-C30, and C20-C38). Its pharmacological target is the Ether-a-gogo potassium channel. We herein determine the solution structure of APETx1 by use of conventional two-dimensional 1H-NMR techniques followed by torsion angle dynamics and refinement protocols. The calculated structure of APETx1 belongs to the disulfide-rich all-beta structural family, in which a three-stranded anti-parallel beta-sheet is the only secondary structure. APETx1 is the first Ether-a-gogo effector discovered to fold in this way. We therefore compare the structure of APETx1 to those of the two other known effectors of the Ether-a-gogo potassium channel, CnErg1 and BeKm-1, and analyze the topological disposition of key functional residues proposed by analysis of the electrostatic anisotropy. The interacting surface is made of a patch of aromatic residues (Y5, Y32, and F33) together with two basic residues (K8 and K18) at the periphery of the surface. We pinpoint the absence of the central lysine present in the functional surface of the two other Ether-a-gogo effectors.
Organizational Affiliation:
Architecture et Fonction des Macromolécules Biologiques, Centre National de la Recherche Scientifique, Unité Mixte de Recherche 6098 and Universités d'Aix-Marseille I and II, Marseille, France.