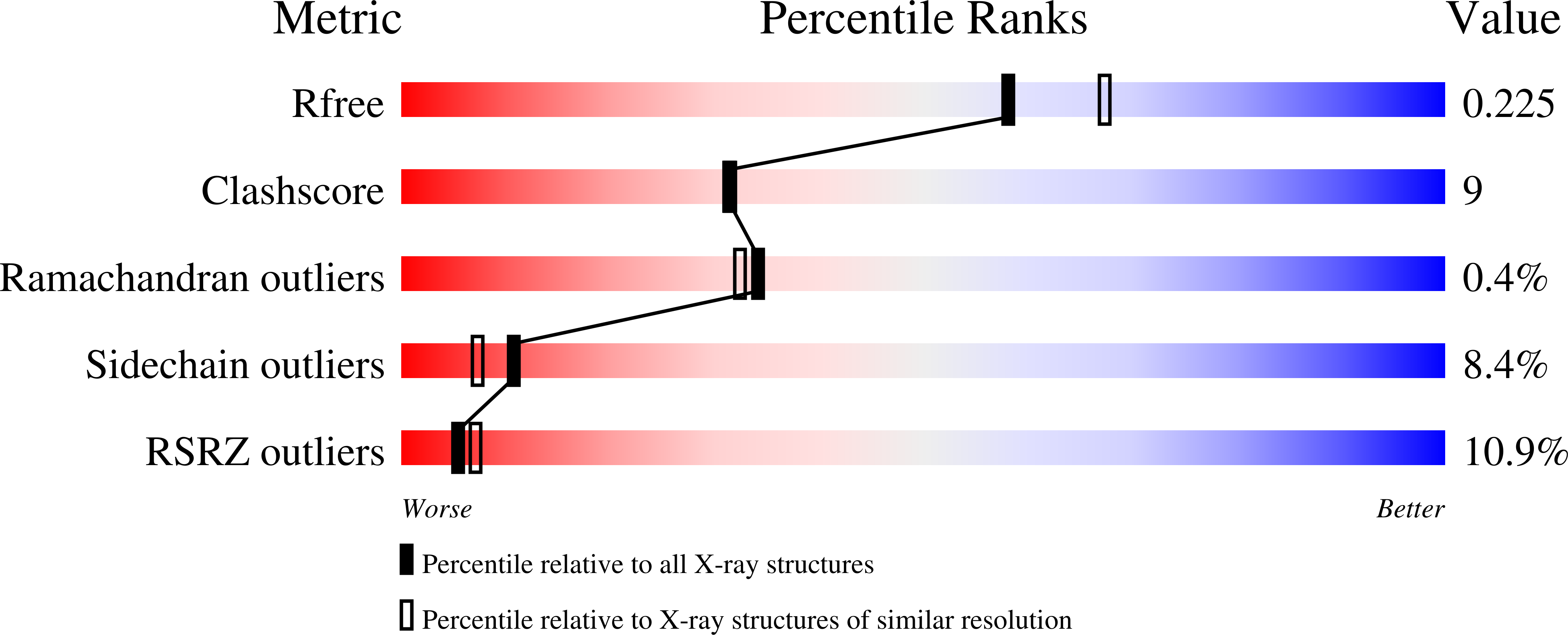
The Skap-hom dimerization and PH domains comprise a 3'-phosphoinositide-gated molecular switch.
Swanson, K.D., Tang, Y., Ceccarelli, D.F., Poy, F., Sliwa, J.P., Neel, B.G., Eck, M.J.(2008) Mol Cell 32: 564-575
- PubMed: 19026786
- DOI: https://doi.org/10.1016/j.molcel.2008.09.022
- Primary Citation of Related Structures:
1U5G, 2OTX - PubMed Abstract:
PH domains, by binding to phosphoinositides, often serve as membrane-targeting modules. Using crystallographic, biochemical, and cell biological approaches, we have uncovered a mechanism that the integrin-signaling adaptor Skap-hom uses to mediate cytoskeletal interactions. Skap-hom is a homodimer containing an N-terminal four-helix bundle dimerization domain, against which its two PH domains pack in a conformation incompatible with phosphoinositide binding. The isolated PH domains bind PI[3,4,5]P(3), and mutations targeting the dimerization domain or the PH domain's PI[3,4,5]P(3)-binding pocket prevent Skap-hom localization to ruffles. Targeting is retained when the PH domain is deleted or by combined mutation of the PI[3,4,5]P(3)-binding pocket and the PH/dimerization domain interface. Thus, the dimerization and PH domain form a PI[3,4,5]P(3)-responsive molecular switch that controls Skap-hom function.
Organizational Affiliation:
Cancer Biology Program, Division of Hematology-Oncology, Department of Medicine, Beth Israel Deaconess Medical Center, Boston, MA 02115, USA.