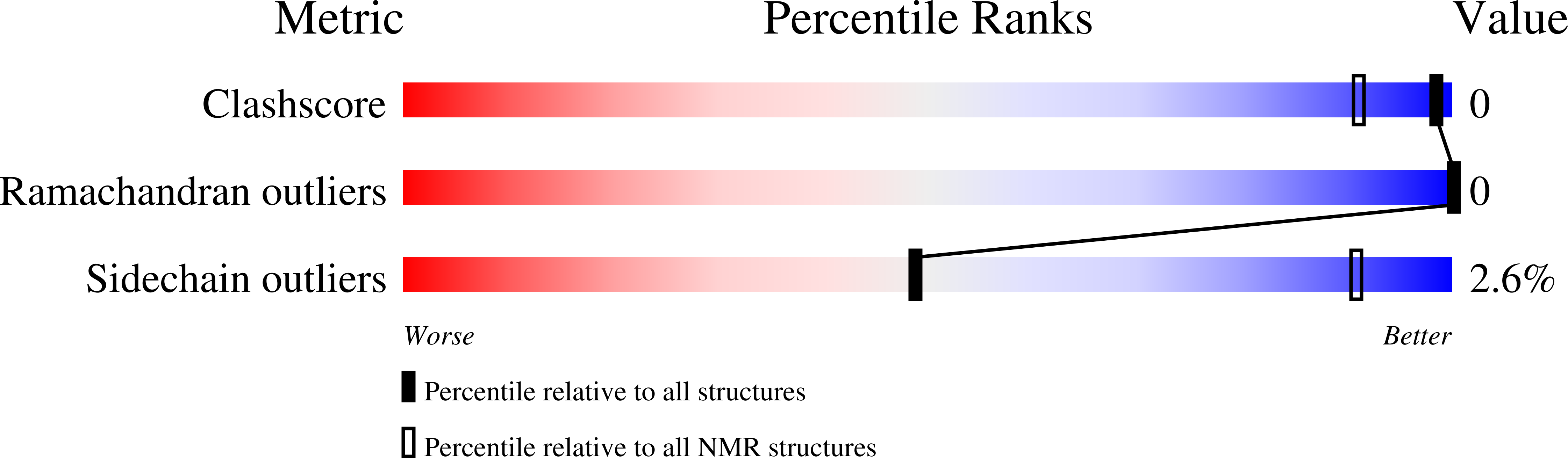Solution Structure of the Symmetric Coiled Coil Tetramer Formed by the Oligomerization Domain of hnRNP C: Implications for Biological Function.
Whitson, S.R., Lestourgeon, W.M., Krezel, A.M.(2005) J Mol Biol 350: 319-337
- PubMed: 15936032
- DOI: https://doi.org/10.1016/j.jmb.2005.05.002
- Primary Citation of Related Structures:
1TXP - PubMed Abstract:
During active cell division, heterogeneous nuclear ribonucleoprotein (hnRNP) C is one of the most abundant proteins in the nucleus. hnRNP C exists as a stable tetramer that binds about 230 nucleotides of pre-mRNA and functions in vivo to package nascent transcripts and nucleate assembly of the 40 S hnRNP complex. Previous studies have shown that monomers lacking or possessing mutant oligomerization domains bind RNA with low affinity, strongly suggesting a cooperative protomer-RNA binding mode. In order to understand the role of the oligomerization domain in defining the biological functions and structure of hnRNP C tetramers, we have determined the high-resolution NMR structure of the oligomerization interface that is formed at the core of the complex, examining specific molecular interactions that drive assembly and contribute to the structural integrity of the tetramer. The determined structure reveals an antiparallel four-helix coiled coil, where classically described knobs-into-holes packing interactions at interhelical contact surfaces are optimized so that side-chains interdigitate to create an even distribution of hydrophobic surfaces along the core. While the stoichiometry of the complex appears to be primarily specified by occlusion of hydrophobic surfaces, particularly the interfacial residue L198, from solvent, helix orientation is primarily determined by electrostatic attractions across helix interfaces. The creation of potential interaction surfaces for other hnRNP C domains along the coiled coil exterior and the assembly of oligomerization interfaces in an antiparallel orientation shape the tertiary fold of full-length monomers and juxtapose RNA-binding elements at distal surfaces of the tetrameric complex in the quaternary assembly. In addition, we discuss the specific challenges encountered in structure determination of this symmetric oligomer by NMR methods, specifically in sorting ambiguous interatomic distance constraints into classes that define different elements of the coiled coil structure.
Organizational Affiliation:
Department of Biological Sciences, 465 21st Ave. South, Vanderbilt University, Nashville, TN 37232, USA.