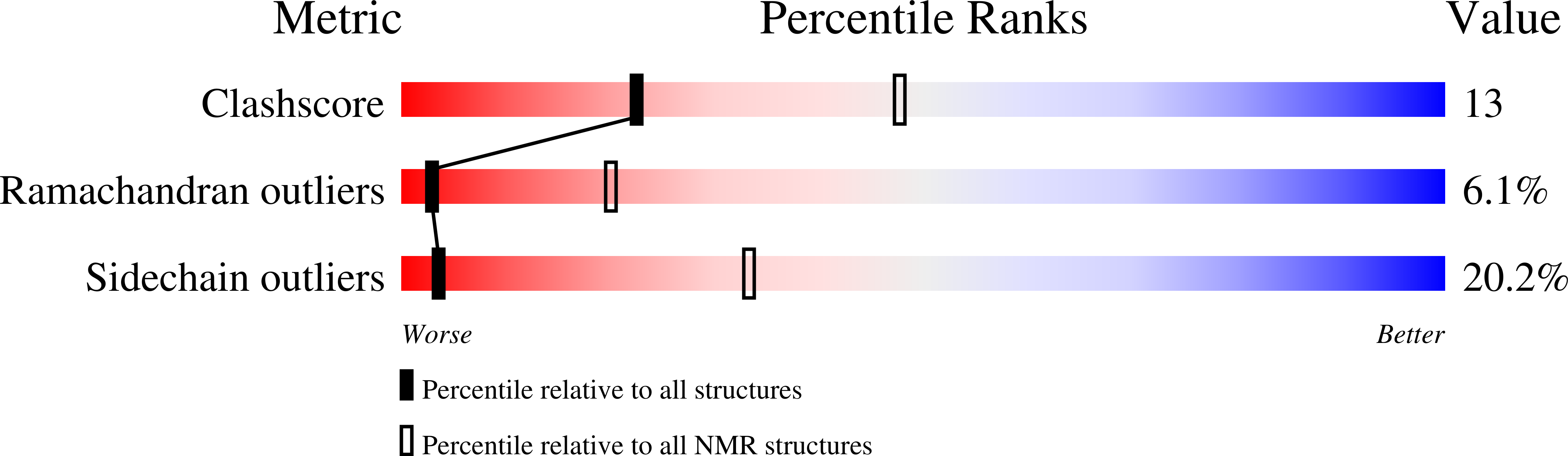Solution structure of maurotoxin, a scorpion toxin from Scorpio maurus, with high affinity for voltage-gated potassium channels.
Blanc, E., Sabatier, J.M., Kharrat, R., Meunier, S., el Ayeb, M., Van Rietschoten, J., Darbon, H.(1997) Proteins 29: 321-333
- PubMed: 9365987
- Primary Citation of Related Structures:
1TXM - PubMed Abstract:
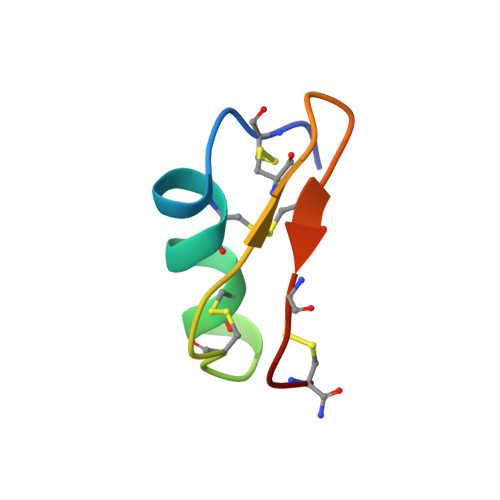
Maurotoxin (MTX), purified from the scorpionid Scorpio maurus is a potent ligand for potassium channels. It shows a broad specificity as being active on Kv1.1 (Kd = 37 nM), Kv1.2 (Kd = 0.8 nM), Kv1.3 (Kd = 150 nM) voltage-gated potassium channels, as well as on small-conductance calcium-activated potassium channels. It has a unique disulfide pairing among the scorpion toxins family. The solution structure of MTX has been determined by 2D-NMR techniques, which led to the full description of its 3D conformation: a bended helix from residues 6 to 16 connected by a loop to a two-stranded antiparallel beta sheet (residues 23 to 26 and 28 to 31). The interaction of MTX with the pore region of the Kv1.2 potassium channel has been modeled according to their charge anisotropy. The structure of MTX is similar to other short scorpion toxins despite its peculiar disulfide pairing. Its interaction with the Kv1.2 channel involves a dipole moment, which guides and orients the toxin onto the pore, toward the binding site, and which thus is responsible for the specificity.
Organizational Affiliation:
AFMB, CNRS UPR 9039, IFR1, Marseille, France.