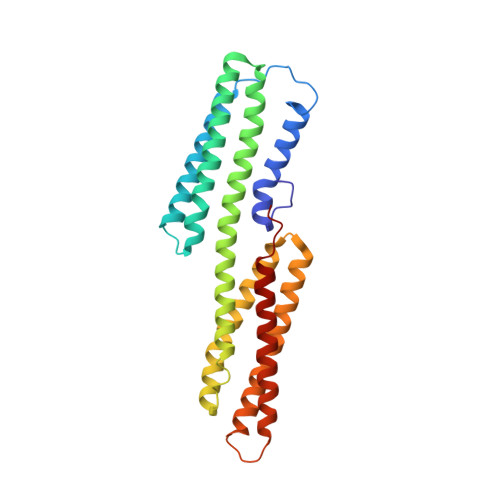
Activation of a vinculin-binding site in the talin rod involves rearrangement of a five-helix bundle
PAPAGRIGORIOU, E., GINGRAS, A.R., BARSUKOV, I.L., Bate, N., Fillingham, I.J., Patel, B., Frank, R., Ziegler, W.H., Roberts, G.C., Critchley, D.R., Emsley, J.(2004) EMBO J 23: 2942-2951
- PubMed: 15272303
- DOI: https://doi.org/10.1038/sj.emboj.7600285
- Primary Citation of Related Structures:
1SJ7, 1SJ8, 1T01 - PubMed Abstract:
The interaction between the cytoskeletal proteins talin and vinculin plays a key role in integrin-mediated cell adhesion and migration. We have determined the crystal structures of two domains from the talin rod spanning residues 482-789. Talin 482-655, which contains a vinculin-binding site (VBS), folds into a five-helix bundle whereas talin 656-789 is a four-helix bundle. We show that the VBS is composed of a hydrophobic surface spanning five turns of helix 4. All the key side chains from the VBS are buried and contribute to the hydrophobic core of the talin 482-655 fold. We demonstrate that the talin 482-655 five-helix bundle represents an inactive conformation, and mutations that disrupt the hydrophobic core or deletion of helix 5 are required to induce an active conformation in which the VBS is exposed. We also report the crystal structure of the N-terminal vinculin head domain in complex with an activated form of talin. Activation of the VBS in talin and the recruitment of vinculin may support the maturation of small integrin/talin complexes into more stable adhesions.
Organizational Affiliation:
Department of Biochemistry, University of Leicester, Leicester, UK.