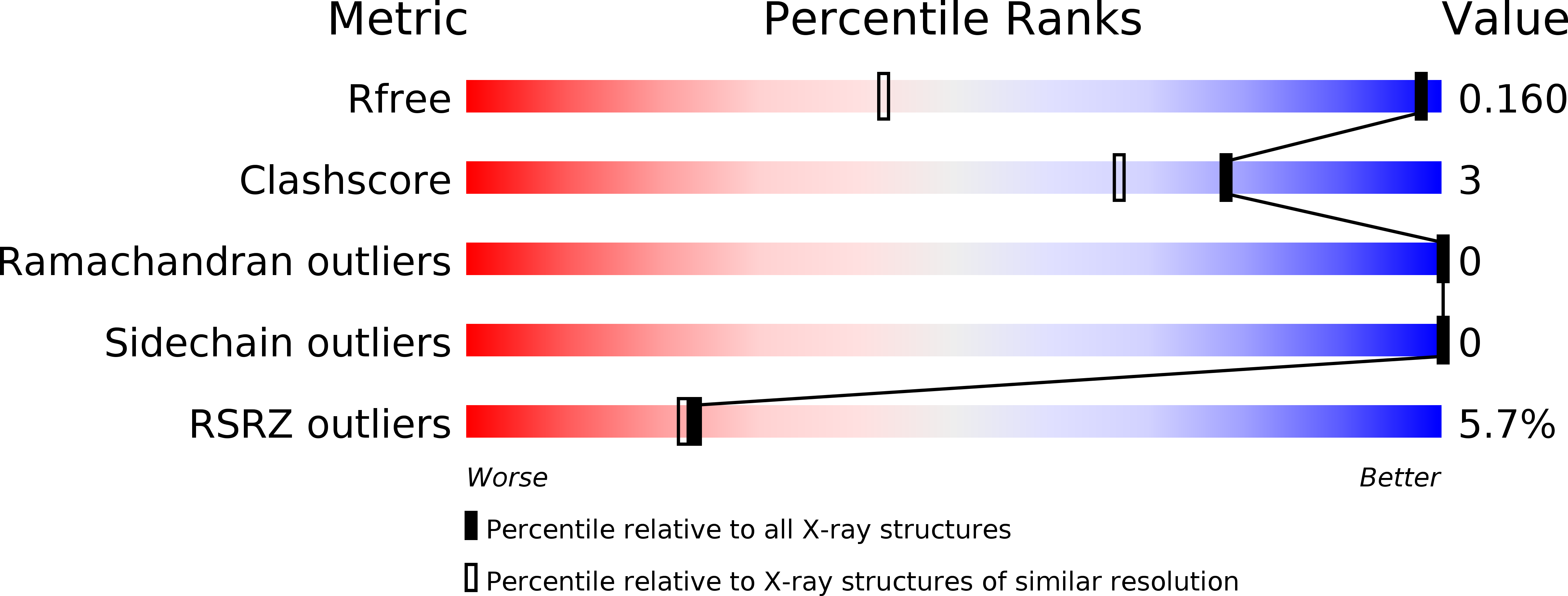
Atomic resolution structure of the HFBII hydrophobin, a self-assembling amphiphile.
Hakanpaa, J., Paananen, A., Askolin, S., Nakari-Setala, T., Parkkinen, T., Penttila, M., Linder, M.B., Rouvinen, J.(2004) J Biol Chem 279: 534-539
- PubMed: 14555650
- DOI: https://doi.org/10.1074/jbc.M309650200
- Primary Citation of Related Structures:
1R2M - PubMed Abstract:
Hydrophobins are proteins specific to filamentous fungi. Hydrophobins have several important roles in fungal physiology, for example, adhesion, formation of protective surface coatings, and the reduction of the surface tension of water, which allows growth of aerial structures. Hydrophobins show remarkable biophysical properties, for example, they are the most powerful surface-active proteins known. To this point the molecular basis of the function of this group of proteins has been largely unknown. We have now determined the crystal structure of the hydrophobin HFBII from Trichoderma reesei at 1.0 A resolution. HFBII has a novel, compact single domain structure containing one alpha-helix and four antiparallel beta-strands that completely envelop two disulfide bridges. The protein surface is mainly hydrophilic, but two beta-hairpin loops contain several conserved aliphatic side chains that form a flat hydrophobic patch that makes the molecule amphiphilic. The amphiphilicity of the HFBII molecule is expected to be a source for surface activity, and we suggest that the behavior of this surfactant is greatly enhanced by the self-assembly that is favored by the combination of size and rigidity. This mechanism of function is supported by atomic force micrographs that show highly ordered arrays of HFBII at the air water interface. The data presented show that much of the current views on structure function relations in hydrophobins must be re-evaluated.
Organizational Affiliation:
Department of Chemistry, University of Joensuu, PO Box 111, 80101 Joensuu, Finland.