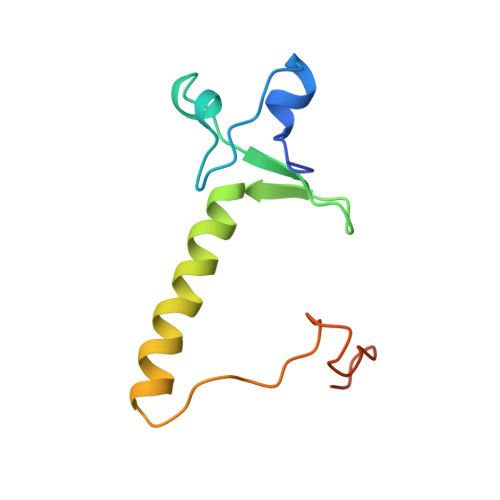
Crystal structure of a PPR1-DNA complex: DNA recognition by proteins containing a Zn2Cys6 binuclear cluster.
Marmorstein, R., Harrison, S.C.(1994) Genes Dev 8: 2504-2512
- PubMed: 7958913
- DOI: https://doi.org/10.1101/gad.8.20.2504
- Primary Citation of Related Structures:
1PYI - PubMed Abstract:
PPR1 is a yeast transcription factor that contains a six-cysteine, two-zinc (Zn) domain, homologous to a similar structure in GAL4. Like GAL4, it binds to DNA sites with conserved CGG triplets symmetrically placed near each end. Whereas the GAL4 site has 11 intervening base pairs, the PPR1 site has 6. The crystal structure of a 95-residue fragment of PPR1 in specific complex with DNA shows that the protein binds to a symmetrical 14-bp recognition site as a nonsymmetrical homodimer. An amino-terminal Zn domain interacts with a conserved CGG triplet near each end of the site through major groove contacts, and the carboxy-terminal residues mediate dimerization through a coiled-coil element and an extended strand. A linker region, connecting the Zn domain and the coiled-coil, folds into a beta-hairpin. This hairpin packs differently on the two subunits and leads to a striking asymmetry, which is largely restricted to the dimerization and linker regions of the protein. Comparison with the GAL4-DNA structure shows that their specificities for sites of different length are determined by the preferred folds of their respective linker segments and by residues at the amino-terminal ends of their coiled-coils. None of these residues contact DNA in PPR1, and they contact only the sugar phosphate backbone in GAL4. We propose that this novel mode of DNA site selection is employed by other proteins that contain a Zn2Cys6 binuclear cluster.
Organizational Affiliation:
Harvard University, Department of Biochemistry and Molecular Biology, Cambridge, Massachusetts.