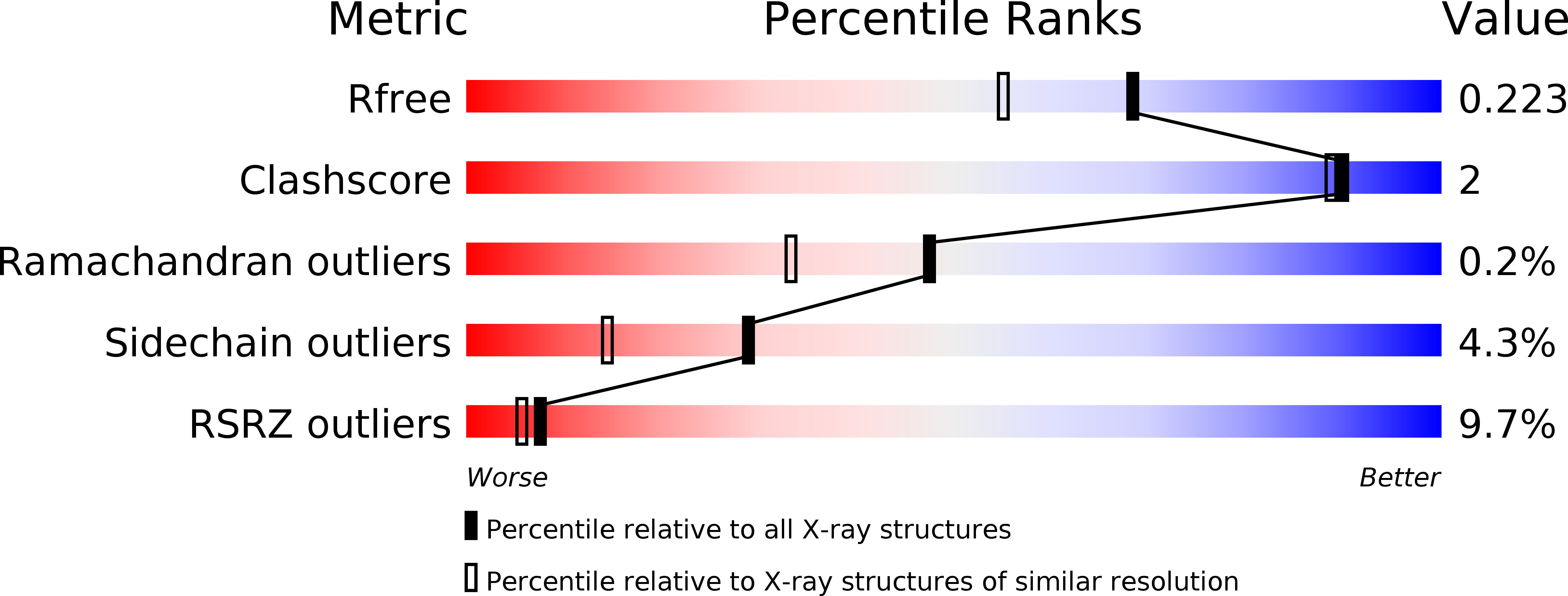
Crystal Structure of Nbla from Anabaena Sp. Pcc 7120, a Small Protein Playing a Key Role in Phycobilisome Degradation.
Bienert, R., Baier, K., Volkmer, R., Lockau, W., Heinemann, U.(2006) J Biol Chem 281: 5216
- PubMed: 16356935
- DOI: https://doi.org/10.1074/jbc.M507243200
- Primary Citation of Related Structures:
1OJH - PubMed Abstract:
Cyanobacterial light-harvesting complexes, the phycobilisomes, are proteolytically degraded when the organisms are starved for combined nitrogen, a process referred to as chlorosis or bleaching. Gene nblA, present in all phycobilisome-containing organisms, encodes a protein of about 7 kDa that plays a key role in phycobilisome degradation. The mode of action of NblA in this degradation process is poorly understood. Here we presented the 1.8-A crystal structure of NblA from Anabaena sp. PCC 7120. In the crystal, NblA is present as a four-helix bundle formed by dimers, the basic structural units. By using pull-down assays with immobilized NblA and peptide scanning, we showed that NblA specifically binds to the alpha-subunits of phycocyanin and phycoerythrocyanin, the main building blocks of the phycobilisome rod structure. By site-directed mutagenesis, we identified amino acid residues in NblA that are involved in phycobilisome binding. The results provided evidence that NblA is directly involved in phycobilisome degradation, and the results allowed us to present a model that gives insight into the interaction of this small protein with the phycobilisomes.
Organizational Affiliation:
Crystallography Group, Max-Delbrück Center for Molecular Medicine, D-13125 Berlin.