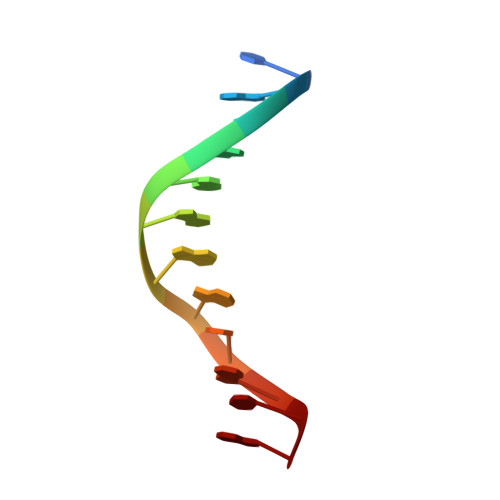
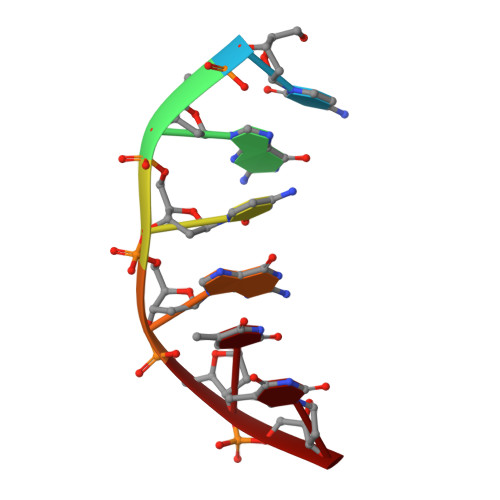
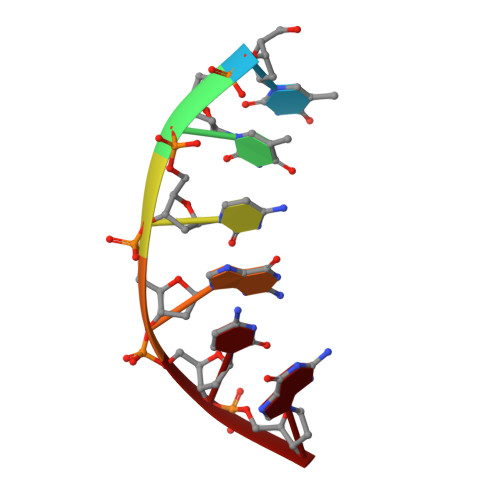
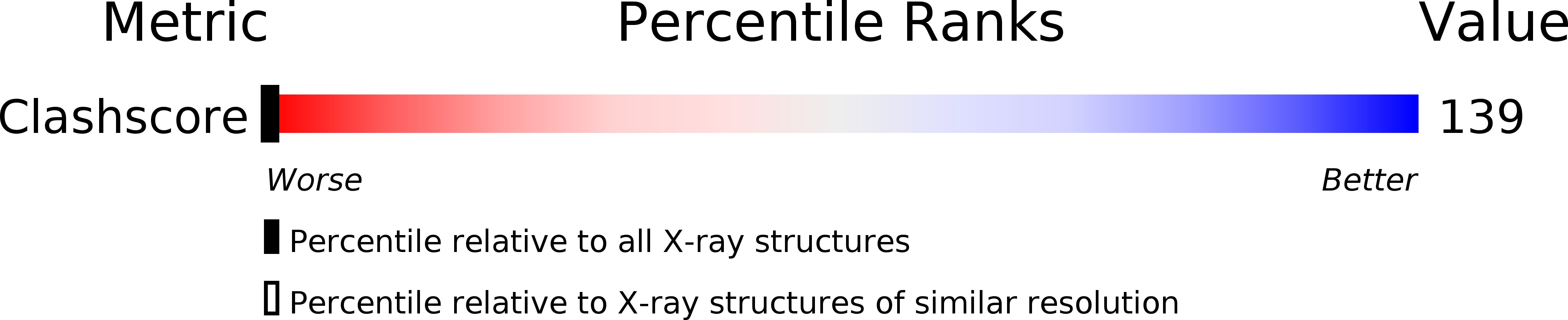Molecular structure of nicked DNA: a substrate for DNA repair enzymes.
Aymami, J., Coll, M., van der Marel, G.A., van Boom, J.H., Wang, A.H., Rich, A.(1990) Proc Natl Acad Sci U S A 87: 2526-2530
- PubMed: 2320572
- DOI: https://doi.org/10.1073/pnas.87.7.2526
- Primary Citation of Related Structures:
1NDN, 1VTE - PubMed Abstract:
The molecular structure of a nicked dodecamer DNA double helix, made of a ternary system containing d(CGCGAAAACGCG) + d(CGCGTT) + d(TTCGCG) oligonucleotides, has been determined by x-ray diffraction analysis at 3 A resolution. The molecule adopts a B-DNA conformation, not unlike those found in intact dodecamer DNA molecules crystallized in a somewhat different crystal lattice, despite a gap due to the absence of a phosphate group in the molecule. The helix has a distinct narrow minor groove near the center of the molecule at the AAAA region. This suggests that the internal stabilizing forces due to base stacking and hydrogen-bonding interactions are sufficient to overcome the loss of connectivity associated with the disruption of the covalent backbone of DNA.
Organizational Affiliation:
Department of Biology, Massachusetts Institute of Technology, Cambridge 02139.