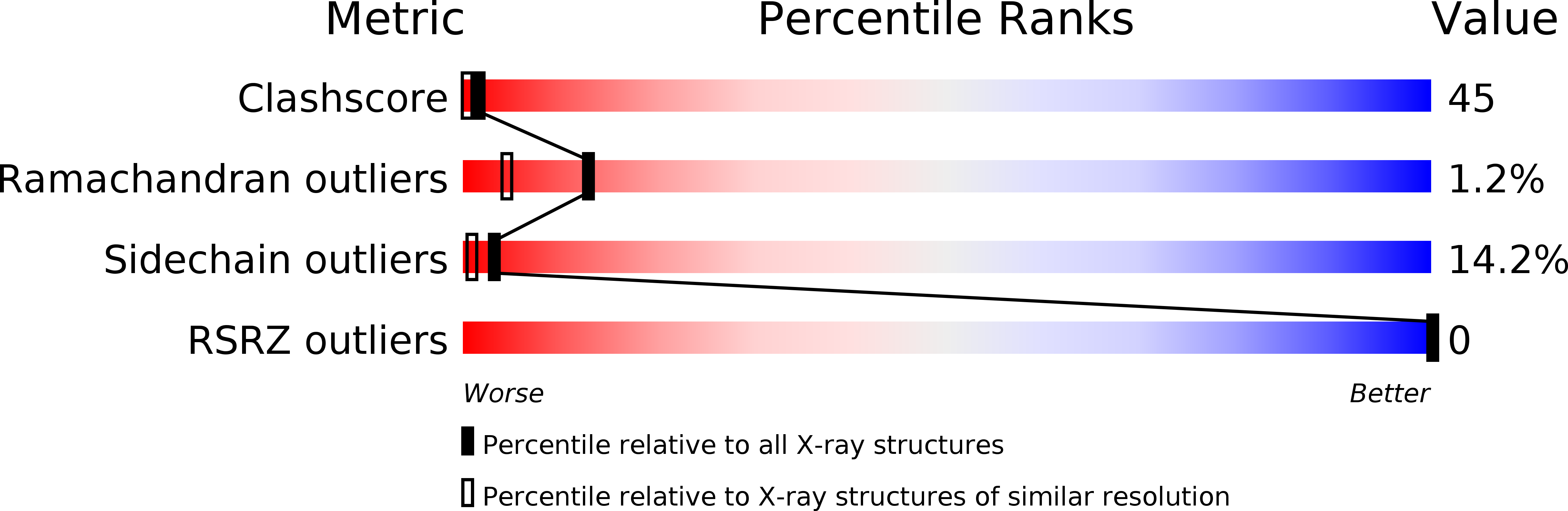
The crystal structure of recombinant human neutrophil-activating peptide-2 (M6L) at 1.9-A resolution.
Malkowski, M.G., Wu, J.Y., Lazar, J.B., Johnson, P.H., Edwards, B.F.(1995) J Biol Chem 270: 7077-7087
- PubMed: 7706245
- DOI: https://doi.org/10.1074/jbc.270.13.7077
- Primary Citation of Related Structures:
1NAP - PubMed Abstract:
Neutrophil-activating peptide-2 (NAP-2) is a 70-residue carboxyl-terminal fragment of platelet basic protein, which is found in the alpha-granules of human platelets. NAP-2, which belongs to the CXC family of chemokines that includes interleukin-8 and platelet factor 4, binds to the interleukin-8 type II receptor and induces a rise in cytosolic calcium, chemotaxis of neutrophils, and exocytosis. Crystals of recombinant NAP-2 in which the single methionine at position 6 was replaced by leucine to facilitate expression belong to space group P1 (unit cell parameters a = 40.8, b = 43.8, and c = 44.7 A and alpha = 98.4 degrees, beta = 120.3 degrees, and gamma = 92.8 degrees), with 4 molecules of NAP-2 (Mr = 7600) in the asymmetric unit. The molecular replacement solution calculated with bovine platelet factor 4 as the starting model was refined using rigid body refinement, manual fitting in solvent-leveled electron density maps, simulated annealing, and restrained least squares to an R-factor of 0.188 for 2 sigma data between 7.0- and 1.9-A resolution. The final refined crystal structure includes 265 solvent molecules. The overall tertiary structure, which is similar to that of platelet factor 4 and interleukin-8, includes an extended amino-terminal loop, three strands of antiparallel beta-sheet arranged in a Greek key fold, and one alpha-helix at the carboxyl terminus. The Glu-Leu-Arg sequence that is critical for receptor binding is fully defined by electron density and exhibits multiple conformations.
Organizational Affiliation:
Department of Biochemistry, Wayne State University, Detroit, Michigan 48201, USA.