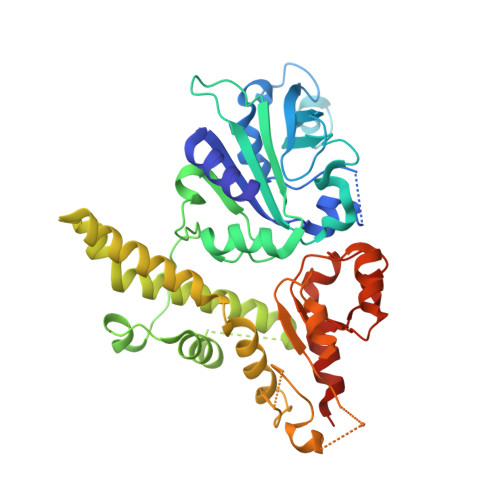
Crystal structure of type IIE restriction endonuclease EcoRII reveals an autoinhibition mechanism by a novel effector-binding fold.
Zhou, X.E., Wang, Y., Reuter, M., Mucke, M., Kruger, D.H., Meehan, E.J., Chen, L.(2004) J Mol Biol 335: 307-319
- PubMed: 14659759
- DOI: https://doi.org/10.1016/j.jmb.2003.10.030
- Primary Citation of Related Structures:
1NA6 - PubMed Abstract:
EcoRII is a type IIE restriction endonuclease that interacts with two copies of the DNA recognition sequence 5'CCWGG, one being the actual target of cleavage, the other serving as the allosteric effector. The mode of enzyme activation by effector binding is unknown. To investigate the molecular basis of activation and cleavage mechanisms by EcoRII, the crystal structure of EcoRII mutant R88A has been solved at 2.1A resolution. The EcoRII monomer has two domains linked through a hinge loop. The N-terminal effector-binding domain has a novel DNA recognition fold with a prominent cleft. The C-terminal catalytic domain has a restriction endonuclease-like fold. Structure-based sequence alignment identified the putative catalytic site of EcoRII that is spatially blocked by the N-terminal domain. The structure together with the earlier characterized EcoRII enzyme activity enhancement in the absence of its N-terminal domain reveal an autoinhibition/activation mechanism of enzyme activity mediated by a novel effector-binding fold. This is the first case of autoinhibition, a mechanism described for many transcription factors and signal transducing proteins, of a restriction endonuclease.
Organizational Affiliation:
Laboratory for Structural Biology, Department of Chemistry, Graduate Programs of Biotechnology, Chemistry and Materials Science, University of Alabama in Huntsville, Huntsville, AL 35899, USA.