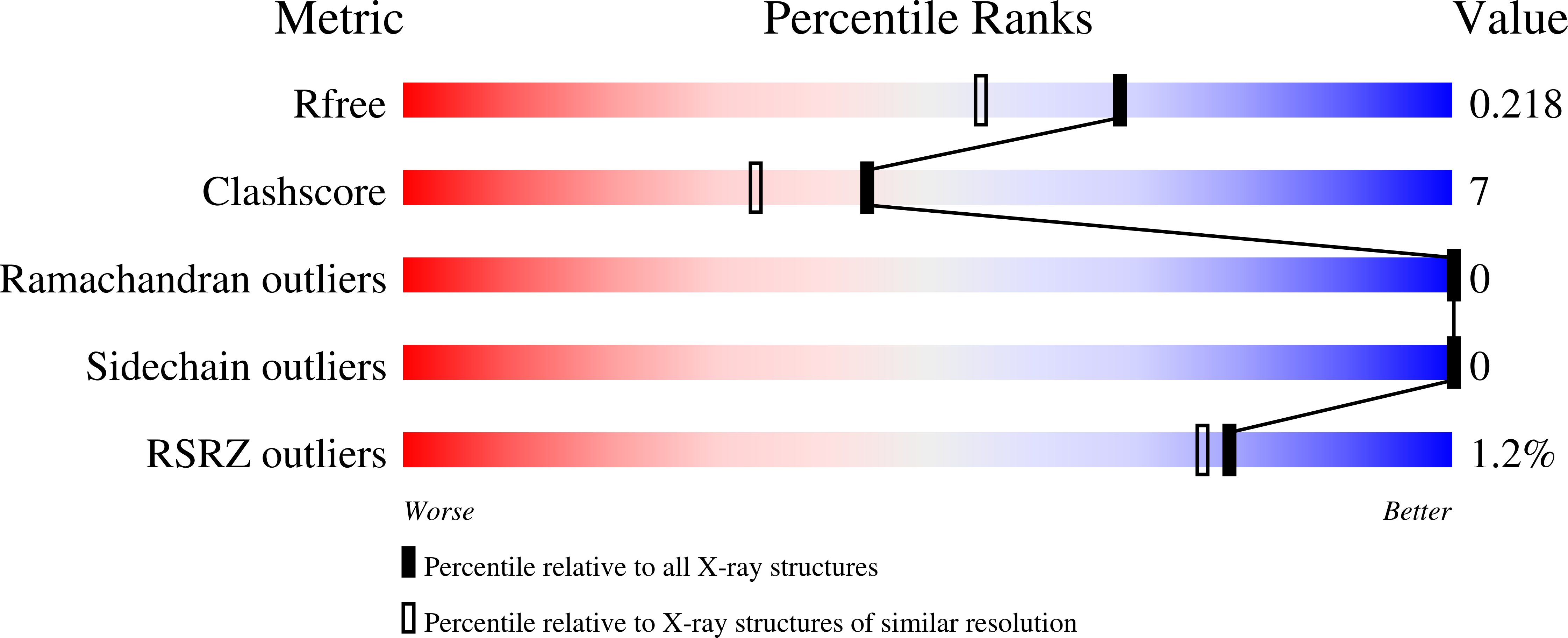
Structures of V45E and V45Y mutants and structure comparison of a variety of cytochrome b5 mutants.
Gan, J.H., Wu, J., Wang, Z.Q., Wang, Y.H., Huang, Z.X., Xia, Z.X.(2002) Acta Crystallogr D Biol Crystallogr 58: 1298-1306
- PubMed: 12136141
- DOI: https://doi.org/10.1107/s0907444902010016
- Primary Citation of Related Structures:
1LQX, 1LR6 - PubMed Abstract:
Val45 is a highly conserved residue and a component of the heme-pocket wall of cytochrome b(5). The crystal structures of cytochrome b(5) mutants V45E and V45Y have been determined at high resolution. Their overall structures were very similar to that of the wild-type protein. However, Val45 of the wild-type protein points towards the heme, but the large side chains of both Glu45 and Tyr45 of the mutants point towards the solvent. A channel is thus opened and the hydrophobicity of the heme pocket is decreased. The rotation of the porphyrin ring and the conformational change of the axial ligand His39 in the V45Y mutant indicate that the microenvironment of the heme is disturbed because of the mutation. The binding constants and the electron-transfer rates between cytochrome b(5) and cytochrome c decrease owing to the mutation, which can be accounted for by molecular modeling: the inter-iron distances increase in order to eliminate the unreasonably close contacts resulting from the large volumes of the mutated side chains. The influence of the mutations on the redox potentials and protein stability is also discussed. The structures of seven mutants of cytochrome b(5) are compared with each other and the effects of these mutations on the protein properties and functions are summarized.
Organizational Affiliation:
State Key Laboratory of Bioorganic and Natural Products Chemistry, Shanghai Institute of Organic Chemistry, Chinese Academy of Sciences, Shanghai 200032, People's Republic of China.