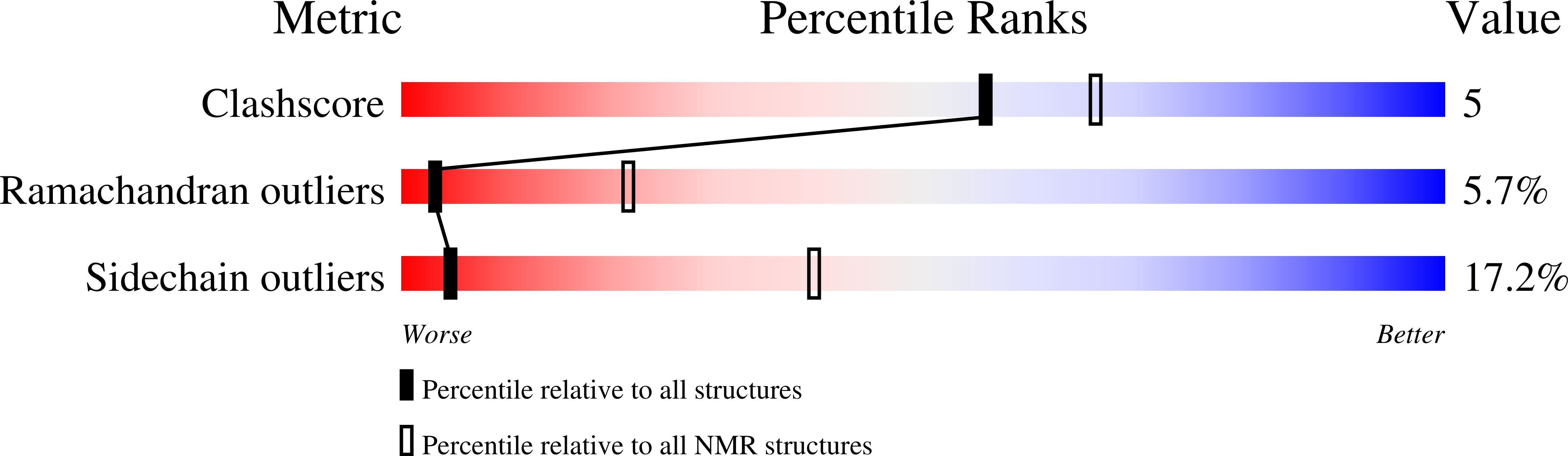Refined structure of lac repressor headpiece (1-56) determined by relaxation matrix calculations from 2D and 3D NOE data: change of tertiary structure upon binding to the lac operator.
Slijper, M., Bonvin, A.M., Boelens, R., Kaptein, R.(1996) J Mol Biol 259: 761-773
- PubMed: 8683581
- DOI: https://doi.org/10.1006/jmbi.1996.0356
- Primary Citation of Related Structures:
1LQC - PubMed Abstract:
The solution structure of the DNA binding domain of lac repressor (headpiece 1-56; HP56) has been refined using data from 2D and 3D NMR spectroscopy. The structure was derived from 1546 restraints (giving an average of 27.6 per residue), comprising 389 intraresidual, 402 sequential, 385 medium range and 325 long range distance restraints and also 30 phi and 15 chi 1 dihedral angle restraints. The structures were determined by the method of direct refinement against nuclear Overhauser enhancement peak volumes with the program DINOSAUR. The final set of 32 selected structures displayed an r.m.s. deviation from the average of 0.43(+/-0.08) A angstroms (backbone) and 0.95(+/-0.08) angstroms (all heavy atoms) for the best defined region of the protein (residues 3 to 49). The ensemble R-factor was 0.35, which indicates close correspondence with the experimental data. The structures revealed good stereochemical qualities. The conformations of the NMR structures of free and DNA complexed lac repressor headpiece were compared. The regions comprising the secondary structure elements show close correspondence for both conformations. However, the conformation of the loop between helix II and III changes considerably upon complexation of the headpiece. This change in the conformation of the loop in lac HP56 is essential for binding of the side-chains of residues Asn25 and His29 to the lac operator DNA. Finally, the lac headpiece residues that are intolerant to mutations were analysed. Most of these mutation-sensitive residues are important for a correct folding of the headpiece region, and a number of these residues are also involved in contacting the operator DNA.
Organizational Affiliation:
Bijvoet Center for Biomolecular Research, Utrecht University, The Netherlands.