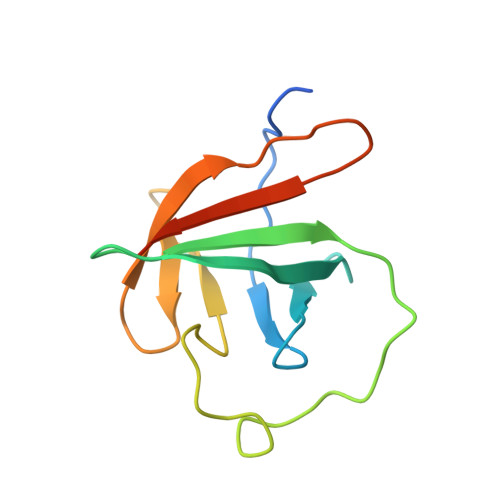
Structure of murine Tcl1 at 2.5 A resolution and implications for the TCL oncogene family.
Petock, J.M., Torshin, I.Y., Wang, Y.F., Du Bois, G.C., Croce, C.M., Harrison, R.W., Weber, I.T.(2001) Acta Crystallogr D Biol Crystallogr 57: 1545-1551
- PubMed: 11679718
- DOI: https://doi.org/10.1107/s090744490101352x
- Primary Citation of Related Structures:
1JNP - PubMed Abstract:
Tcl1 and Mtcp1, members of the Tcl1 family, are implicated in T-cell prolymphocytic leukemia. The crystal structure of a dimer of murine Tcl1 has been determined at 2.5 A resolution with an R factor of 0.225. Murine Tcl1, human Tcl1 and Mtcp1 share very similar subunit structures, with RMS differences of 0.6 and 1.4 A for C(alpha) atoms, respectively, while the sequences share 50 and 36% identity, respectively. These structures fold into an eight-stranded beta-barrel of unique topology and high internal symmetry of 1.1-1.3 A for the two halves of human and murine Tcl1 and 1.7 A for Mtcp1, despite the low 12-13% sequence identity. The molecular surfaces of all three structures showed a common planar region which is likely to be involved in protein-protein interactions.
Organizational Affiliation:
Department of Biology, Georgia State University, Atlanta, GA 30303, USA.