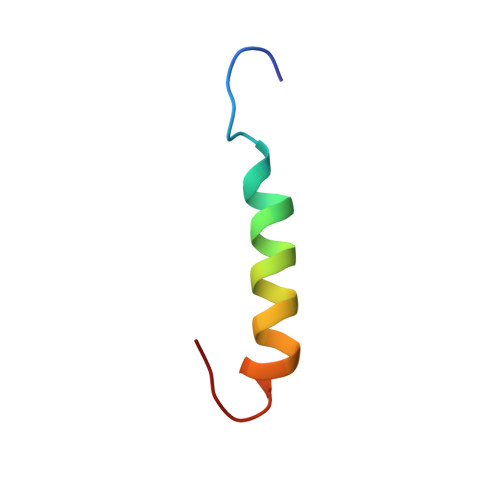
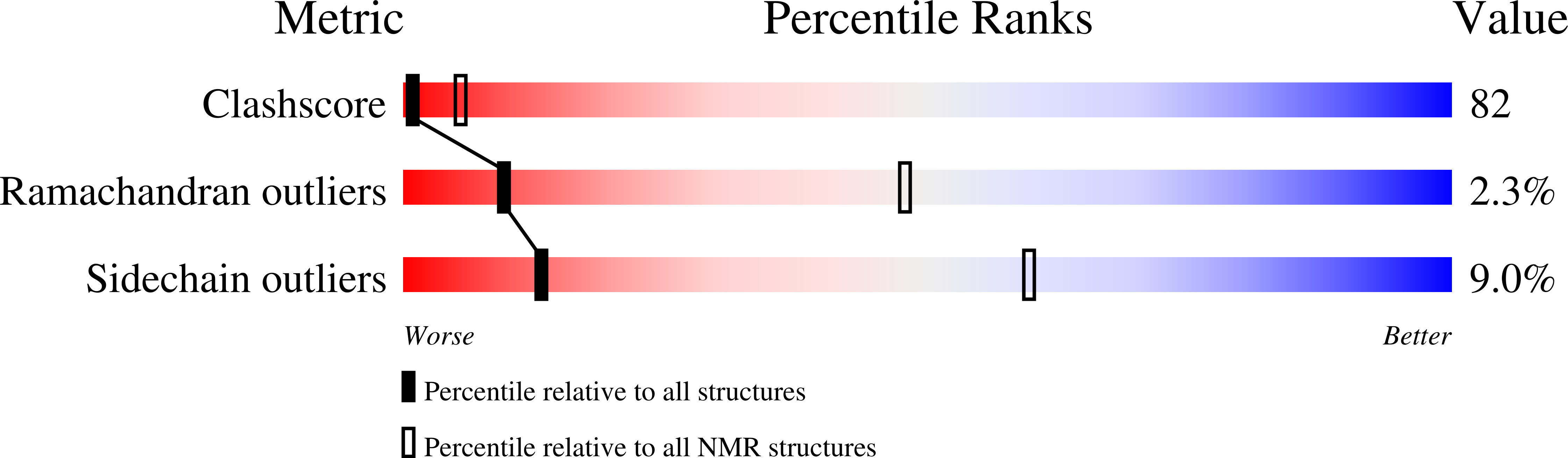Structure and orientation of sarcolipin in lipid environments.
Mascioni, A., Karim, C., Barany, G., Thomas, D.D., Veglia, G.(2002) Biochemistry 41: 475-482
- PubMed: 11781085
- DOI: https://doi.org/10.1021/bi011243m
- Primary Citation of Related Structures:
1JDM - PubMed Abstract:
Sarcolipin (SLN) is a 31 amino acid integral membrane protein that regulates Ca-ATPase activity in skeletal muscle. Here, we report the three-dimensional structure and topology of synthetic SLN in lipid environments, as determined by solution and solid-state NMR spectroscopy. 2D solution NMR experiments were performed on SLN solubilized in sodium dodecyl sulfate (SDS) micelles. We found that SLN adopts a highly defined alpha-helical conformation from F9 through R27, with a backbone RMSD of 0.65 A and a side chain RMSD of 1.66 A. The N-terminus (M1 through L8) and the C-terminus (S28 through Y31) are mostly unstructured. The orientation of the SLN was determined using one-dimensional (15)N NMR solid-state spectroscopy. The protein was incorporated into phospholipid bilayers prepared from a mixture of 1,2-dioleoyl-sn-glycero-3-phosphocholine and 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine. The (15)N chemical shift solid-state spectra from selectively labeled SLN samples indicate that SLN orients perpendicularly to the plane of the membrane bilayers. These results support the proposed mechanism of Ca-ATPase regulation of SLN via protein-protein intramembranous interactions between the highly conserved transmembrane domains of the Ca-ATPase and the conserved transmembrane domain of SLN.
Organizational Affiliation:
Department of Chemistry, University of Minnesota, Minneapolis, Minnesota 55455, USA.