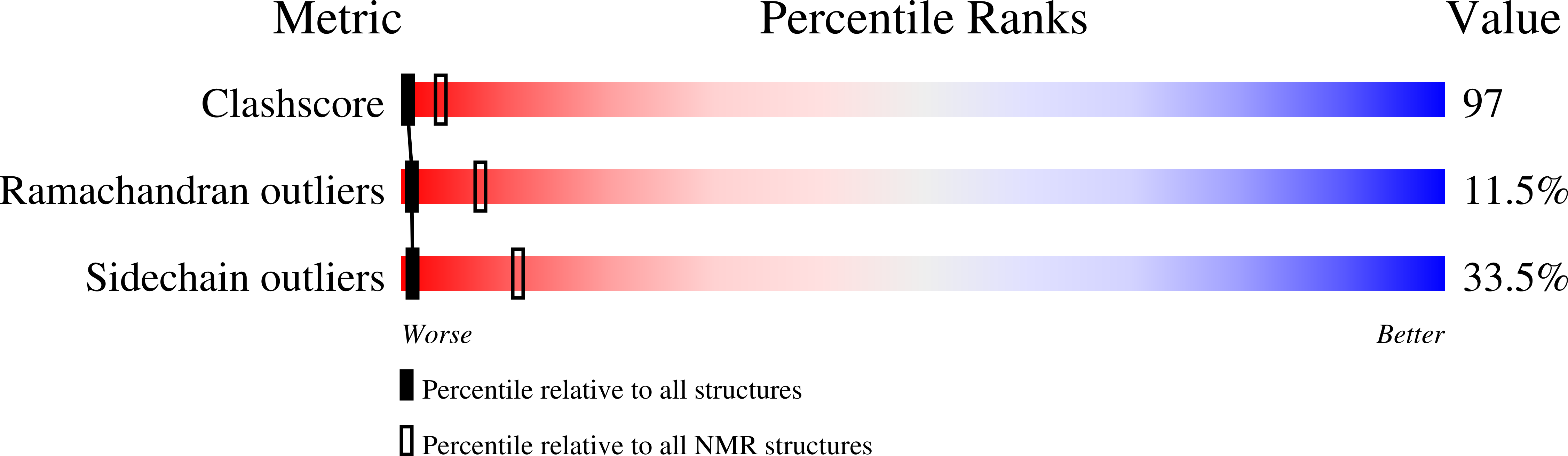The S. cerevisiae architectural HMGB protein NHP6A complexed with DNA: DNA and protein conformational changes upon binding
Masse, J.E., Wong, B., Yen, Y.-M., Allain, F.H.-T., Johnson, R.C., Feigon, J.(2002) J Mol Biol 323: 263-284
- PubMed: 12381320
- DOI: https://doi.org/10.1016/s0022-2836(02)00938-5
- Primary Citation of Related Structures:
1J5N, 1LWA, 1LWM - PubMed Abstract:
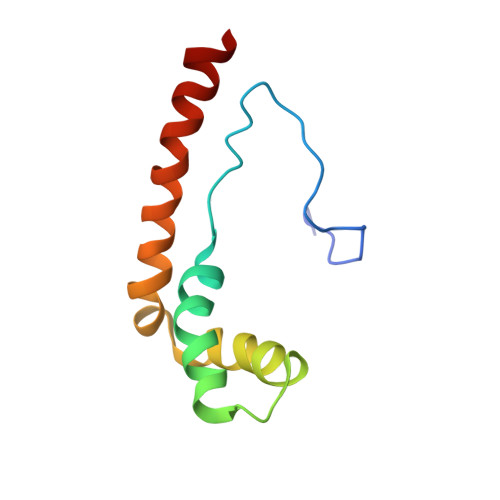
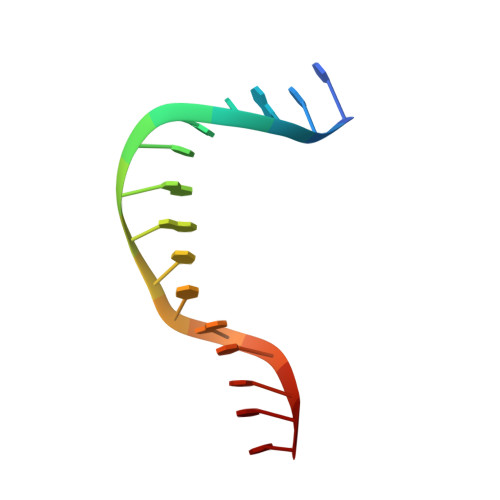
NHP6A is a non-sequence-specific DNA-binding protein from Saccharomyces cerevisiae which belongs to the HMGB protein family. Previously, we have solved the structure of NHP6A in the absence of DNA and modeled its interaction with DNA. Here, we present the refined solution structures of the NHP6A-DNA complex as well as the free 15bp DNA. Both the free and bound forms of the protein adopt the typical L-shaped HMGB domain fold. The DNA in the complex undergoes significant structural rearrangement from its free form while the protein shows smaller but significant conformational changes in the complex. Structural and mutational analysis as well as comparison of the complex with the free DNA provides insight into the factors that contribute to binding site selection and DNA deformations in the complex. Further insight into the amino acid determinants of DNA binding by HMGB domain proteins is given by a correlation study of NHP6A and 32 other HMGB domains belonging to both the DNA-sequence-specific and non-sequence-specific families of HMGB proteins. The resulting correlations can be rationalized by comparison of solved structures of HMGB proteins.
Organizational Affiliation:
Department of Chemistry and Biochemistry, University of California at Los Angeles, Los Angeles, CA 90095-1569, USA.