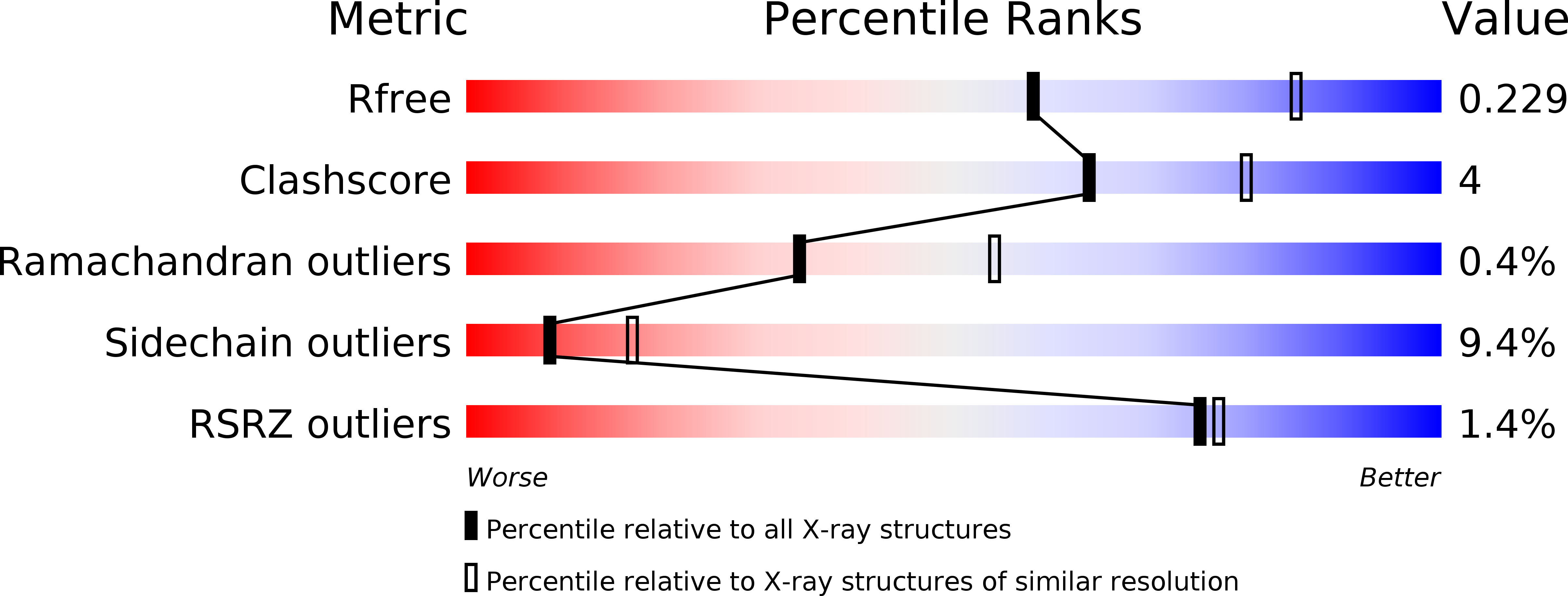
The crystal structure of indoleglycerol-phosphate synthase from Thermotoga maritima. Kinetic stabilization by salt bridges.
Knochel, T., Pappenberger, A., Jansonius, J.N., Kirschner, K.(2002) J Biol Chem 277: 8626-8634
- PubMed: 11741953
- DOI: https://doi.org/10.1074/jbc.M109517200
- Primary Citation of Related Structures:
1I4N - PubMed Abstract:
The crystal structure of the thermostable indoleglycerol-phosphate synthase from Thermotoga maritima (tIGPS) was determined at 2.5 A resolution. It was compared with the structures of the thermostable sIGPS from Sulfolobus solfataricus and of the thermolabile eIGPS from Escherichia coli. The main chains of the three (beta alpha)(8)-barrel proteins superimpose closely, and the packing of side chains in the beta-barrel cores, as well as the architecture of surface loops, is very similar. Both thermostable proteins have, however, 17 strong salt bridges, compared with only 10 in eIGPS. The number of additional salt bridges in tIGPS and sIGPS correlates well with their reduced rate of irreversible thermal inactivation at 90 degrees C. Only 3 of 17 salt bridges in tIGPS and sIGPS are topologically conserved. The major difference between the two proteins is the preference for interhelical salt bridges in sIGPS and intrahelical ones in tIGPS. The different implementation of salt bridges in the closely related proteins suggests that the stabilizing effect of salt bridges depends rather on the sum of their individual contributions than on their location. This observation is consistent with a protein unfolding mechanism where the simultaneous breakdown of all salt bridges is the rate-determining step.
Organizational Affiliation:
Division of Structural Biology, Biozentrum, University Basel, Klingelbergstrasse 70, CH-4056 Basel, Switzerland. thorsten_knoechel@sandwich.pfizer.com