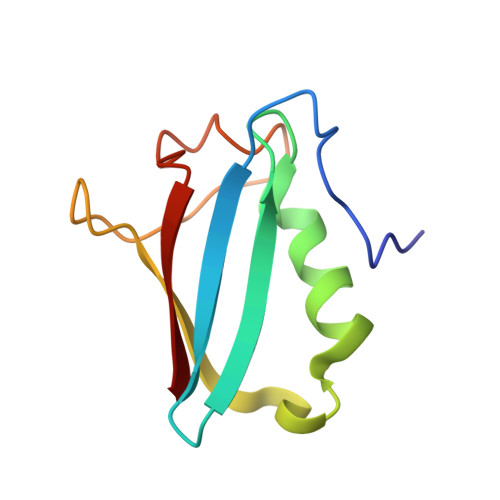
Solution structure and interaction surface of the C-terminal domain from p47: a major p97-cofactor involved in SNARE disassembly.
Yuan, X., Shaw, A., Zhang, X., Kondo, H., Lally, J., Freemont, P.S., Matthews, S.(2001) J Mol Biol 311: 255-263
- PubMed: 11478859
- DOI: https://doi.org/10.1006/jmbi.2001.4864
- Primary Citation of Related Structures:
1I42, 1JRU - PubMed Abstract:
p47 is the major protein identified in complex with the cytosolic AAA ATPase p97. It functions as an essential cofactor of p97-regulated membrane fusion, which has been suggested to disassemble t-t-SNARE complexes and prepare them for further rounds of membrane fusion. Here, we report the high-resolution NMR structure of the C-terminal domain from p47. It comprises a UBX domain and a 13 residue long structured N-terminal extension. The UBX domain adopts a characteristic ubiquitin fold with a betabetaalphabetabetaalphabeta secondary structure arrangement. Three hydrophobic residues from the N-terminal extension pack closely against a cleft in the UBX domain. We also identify, for the first time, the p97 interaction surface using NMR chemical shift perturbation studies.
Organizational Affiliation:
Department of Biological Sciences, Wolfson Laboratories, Imperial College of Science Technology and Medicine, London, South Kensington, SW7 2AY, UK.