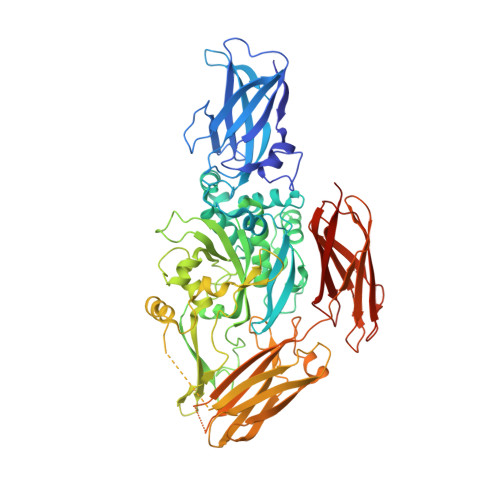
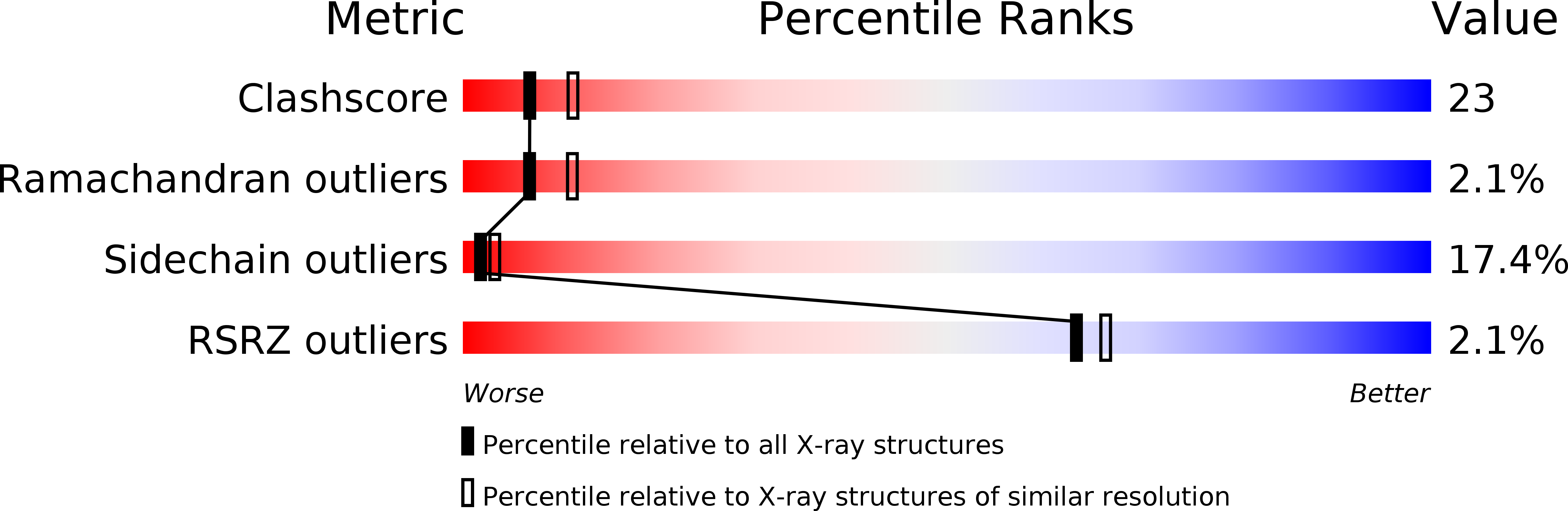Crystal structure of red sea bream transglutaminase.
Noguchi, K., Ishikawa, K., Yokoyama, K.i., Ohtsuka, T., Nio, N., Suzuki, E.(2001) J Biol Chem 276: 12055-12059
- PubMed: 11080504
- DOI: https://doi.org/10.1074/jbc.M009862200
- Primary Citation of Related Structures:
1G0D - PubMed Abstract:
The crystal structure of the tissue-type transglutaminase from red sea bream liver (fish-derived transglutaminase, FTG) has been determined at 2.5-A resolution using the molecular replacement method, based on the crystal structure of human blood coagulation factor XIII, which is a transglutaminase zymogen. The model contains 666 residues of a total of 695 residues, 382 water molecules, and 1 sulfate ion. FTG consists of four domains, and its overall and active site structures are similar to those of human factor XIII. However, significant structural differences are observed in both the acyl donor and acyl acceptor binding sites, which account for the difference in substrate preferences. The active site of the enzyme is inaccessible to the solvent, because the catalytic Cys-272 hydrogen-bonds to Tyr-515, which is thought to be displaced upon acyl donor binding to FTG. It is postulated that the binding of an inappropriate substrate to FTG would lead to inactivation of the enzyme because of the formation of a new disulfide bridge between Cys-272 and the adjacent Cys-333 immediately after the displacement of Tyr-515. Considering the mutational studies previously reported on the tissue-type transglutaminases, we propose that Cys-333 and Tyr-515 are important in strictly controlling the enzymatic activity of FTG.
Organizational Affiliation:
Central Research Laboratories and Food Research & Development Laboratories, Ajinomoto Company Inc., 1-1 Suzuki-cho, Kawasaki-ku, Kawasaki, Kanagawa 210-8681, Japan.