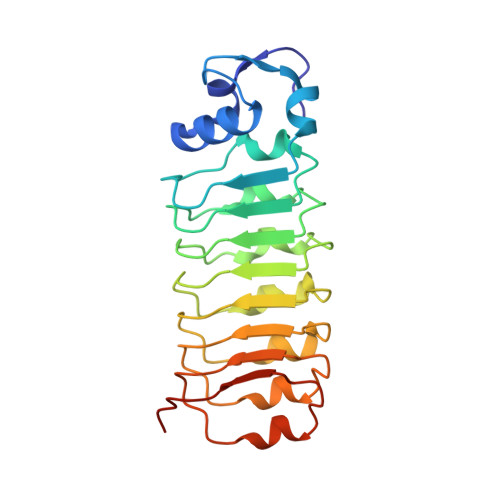
Structure of the lnlB leucine-rich repeats, a domain that triggers host cell invasion by the bacterial pathogen L. monocytogenes.
Marino, M., Braun, L., Cossart, P., Ghosh, P.(1999) Mol Cell 4: 1063-1072
- PubMed: 10635330
- DOI: https://doi.org/10.1016/s1097-2765(00)80234-8
- Primary Citation of Related Structures:
1D0B - PubMed Abstract:
The L. monocytogenes protein lnlB activates phosphoinositide 3-kinase and induces phagocytosis in several mammalian cell types. The 1.86 A resolution X-ray crystal structure of the leucine-rich repeat domain of lnlB that is both necessary and sufficient to induce phagocytosis is presented here. The structure supports a crucial role for calcium in host cell invasion by L. monocytogenes and supplies a rationale for its function. Calciums are bound to the protein in an unusually exposed manner that suggests that the metals may act as a bridge between lnlB and mammalian cell surface receptors. The structure also identifies surfaces on the curved and elongated molecule that may constitute additional interaction sites in forming a bacterial-mammalian signaling complex.
Organizational Affiliation:
Department of Chemistry and Biochemistry, University of California, San Diego, La Jolla 92093-0314, USA.