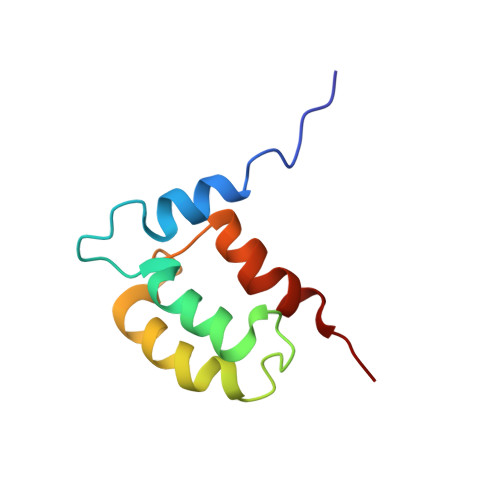
Solution structure and backbone dynamics of Calsensin, an invertebrate neuronal calcium-binding protein.
Venkitaramani, D.V., Fulton, D.B., Andreotti, A.H., Johansen, K.M., Johansen, J.(2005) Protein Sci 14: 1894-1901
- PubMed: 15937283
- DOI: https://doi.org/10.1110/ps.051412605
- Primary Citation of Related Structures:
1YX7, 1YX8 - PubMed Abstract:
Calsensin is an EF-hand calcium-binding protein expressed by a subset of peripheral sensory neurons that fasciculate into a single tract in the leech central nervous system. Calsensin is a 9-kD protein with two EF-hand calcium-binding motifs. Using multidimensional NMR spectroscopy we have determined the solution structure and backbone dynamics of calcium-bound Calsensin. Calsensin consists of four helices forming a unicornate-type four-helix bundle. The residues in the third helix undergo slow conformational exchange indicating that the motion of this helix is associated with calciumbinding. The backbone dynamics of the protein as measured by (15)N relaxation rates and heteronuclear NOEs correlate well with the three-dimensional structure. Furthermore, comparison of the structure of Calsensin with other members of the EF-hand calcium-binding protein family provides insight into plausible mechanisms of calcium and target protein binding.
Organizational Affiliation:
Department of Biochemistry, Biophysics, and Molecular Biology, Iowa State University, Ames, IA 50011, USA.