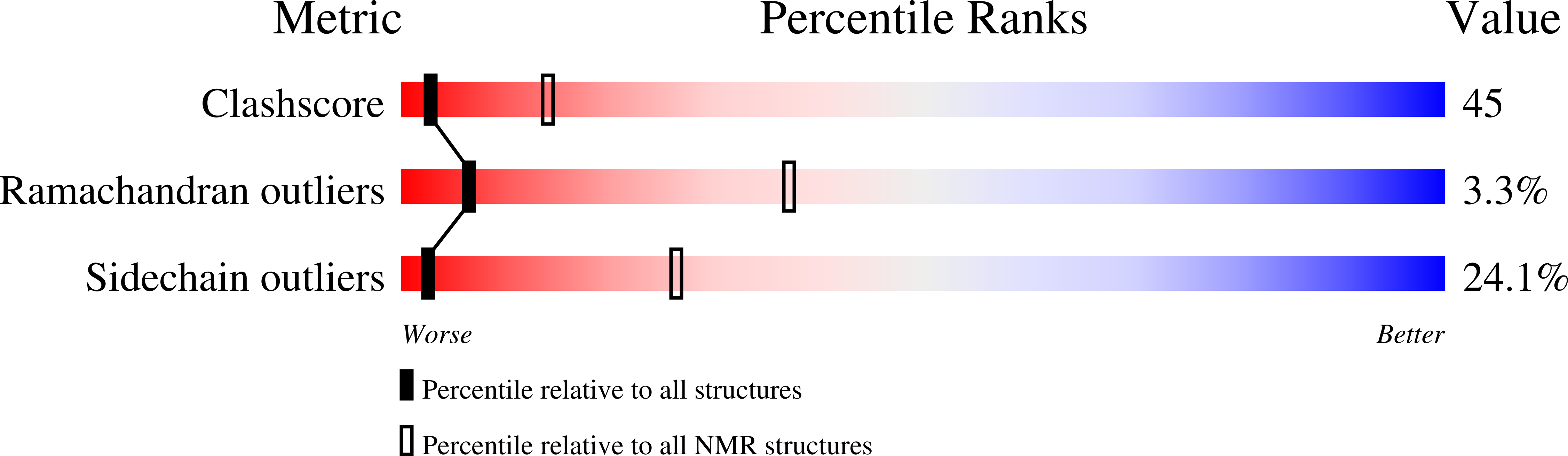Structure and Function of a Bacterial Fasciclin I Domain Protein Elucidates Function of Related Cell Adhesion Proteins Such as Tgfbip and Periostin.
Moody, R.G., Williamson, M.P.(2013) FEBS Open Bio 3: 71
- PubMed: 23772377
- DOI: https://doi.org/10.1016/j.fob.2013.01.001
- Primary Citation of Related Structures:
1W7D, 1W7E - PubMed Abstract:
Fasciclin I (FAS1) domains have important roles in cell adhesion, which are not understood despite many structural and functional studies. Examples of FAS1 domain proteins include TGFBIp (βig-h3) and periostin, which function in angiogenesis and development of cornea and bone, and are also highly expressed in cancer tissues. Here we report the structure of a single-domain bacterial fasciclin I protein, Fdp, in the free-living photosynthetic bacterium Rhodobacter sphaeroides, and show that it confers cell adhesion properties in vivo. A binding site is identified which includes the most highly conserved region and is adjacent to the N-terminus. By mapping this onto eukaryotic homologues, which all contain tandem FAS1 domains, it is concluded that the interaction site is normally buried in the dimer interface. This explains why corneal dystrophy mutations are concentrated in the C-terminal domain of TGFBIp and suggests new therapeutic approaches.
Organizational Affiliation:
Dept. of Molecular Biology and Biotechnology, Firth Court, Western Bank, University of Sheffield, Sheffield S10 2TN, UK.